Discussion
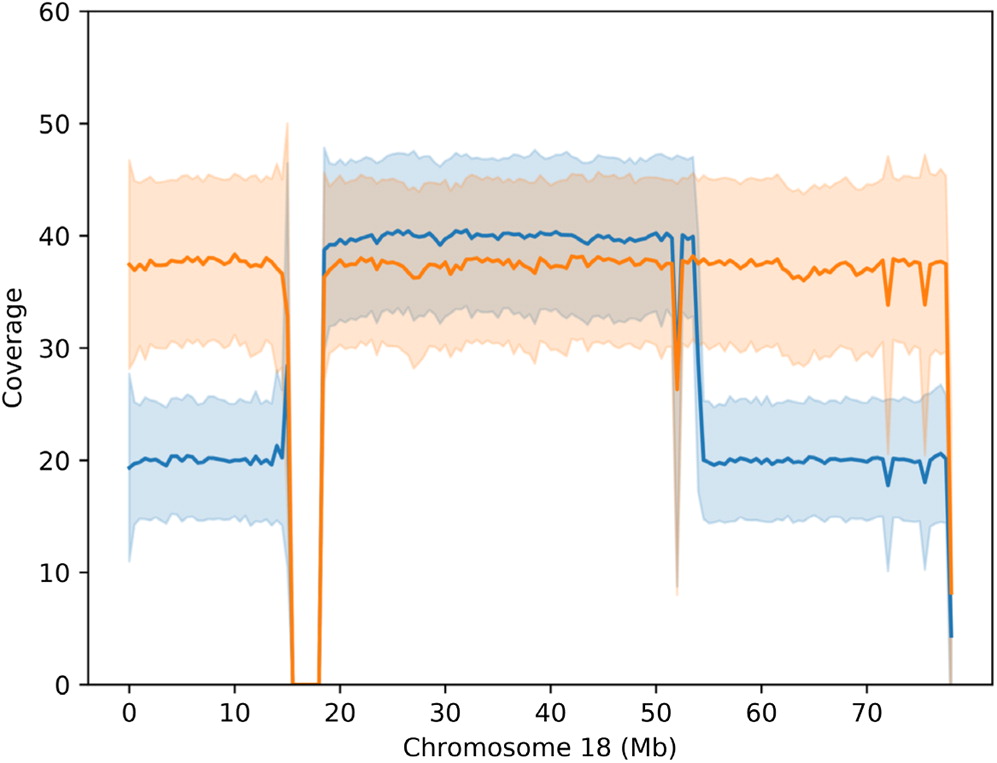
The phenotype of individuals with a given ring chromosome can simply resemble those with terminal deletions, but without ring formation. Nevertheless, given the secondary genomic instability, clinical presentations can be highly variable (
Guilherme et al. 2011). Chromosome 18 was among the first chromosomes found in humans to be affected by ring formation (
Carter et al. 2015). Although deletions of 2 regions on 18q (17 000 000–19 667 062 and 45 578 734–46 739 965) are reportedly fatal, large deletions of chromosome 18 are thought to be less lethal due to the low gene density per Mb (
Cody et al. 2015). Deletions of 18q are thought to have a more variable and unpredictable phenotype compared to 18p deletions. This is likely due to the high variability of hemizygosities as well as other factors such as duplications, somatic mosaicism and ring instability (
Cody et al. 2009;
Carter et al. 2015;
Hasi-Zogaj et al. 2015). Our patient also has a 500 kb hemizygous deletion on chromosome 13 that does not encompass any UCSC genes.
13% of patients with 18p- have low serum immunoglobulin levels. SIgAD is the most common immune defect in any type of chromosome 18 abnormality (
Cody et al. 2015;
Hasi-Zogaj et al. 2015). Distal hemizygosity of 18q22.3-q23 has been also shown to be associated with SIgAD (
Dostal et al. 2007). However, normal serum IgA levels in our patient could be due the variable penetrance of this locus that has been estimated to be between 33% and 50% (
Cody et al. 2015). Additionally, the fact that SIgAD also affects patients with 18p-, suggests that more than 1 single locus on chromosome 18 regulates IgA. Indeed, there is evidence that there is a common genetic basis for SIgAD and CVID as the occurrence of both diseases in the same family or progression of SIgAD to CVID has been observed (
Aghamohammadi et al. 2008). The conversion of SIgAD to CVID has also been previously reported in a female patient with 18q-syndrome (
Slyper and Pietryga 1997). Linkage analysis has failed to show any loci on chromosome 18 associated with SIgAD (
Vorechovsky et al. 1999). Nonetheless, given the technical limitations at the time of those earlier studies, a re-assessment of patients using recently developed technologies such as arrays or next generation sequencing might help to discover novels gene defects or genomic variations implicated in SIgAD or CVID.
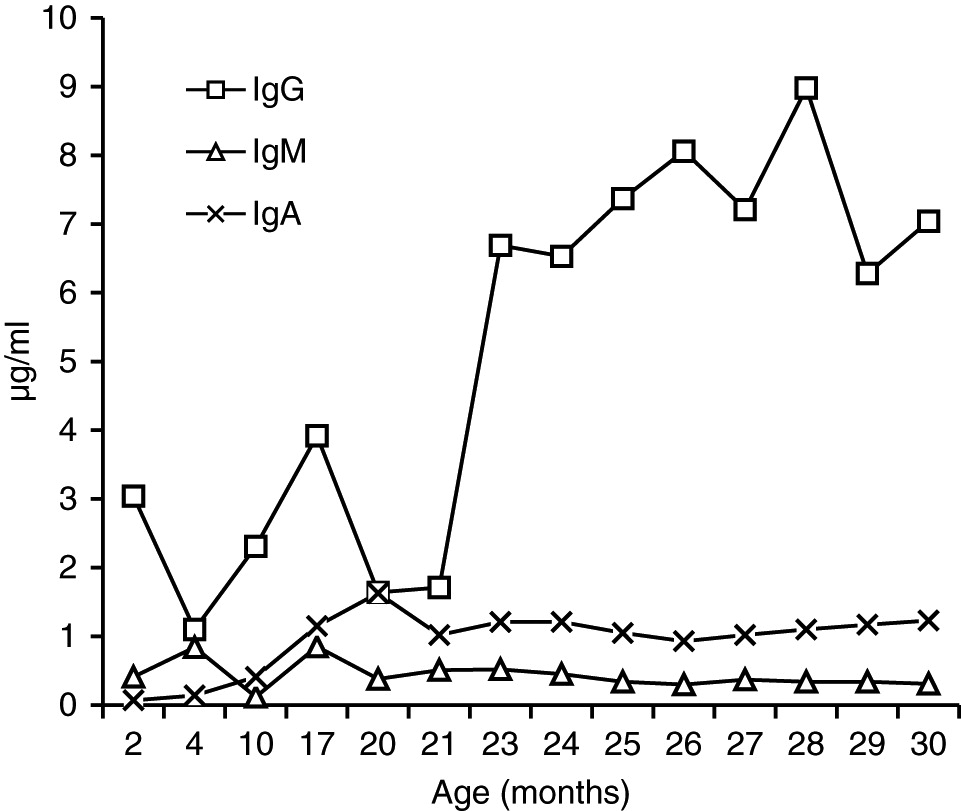
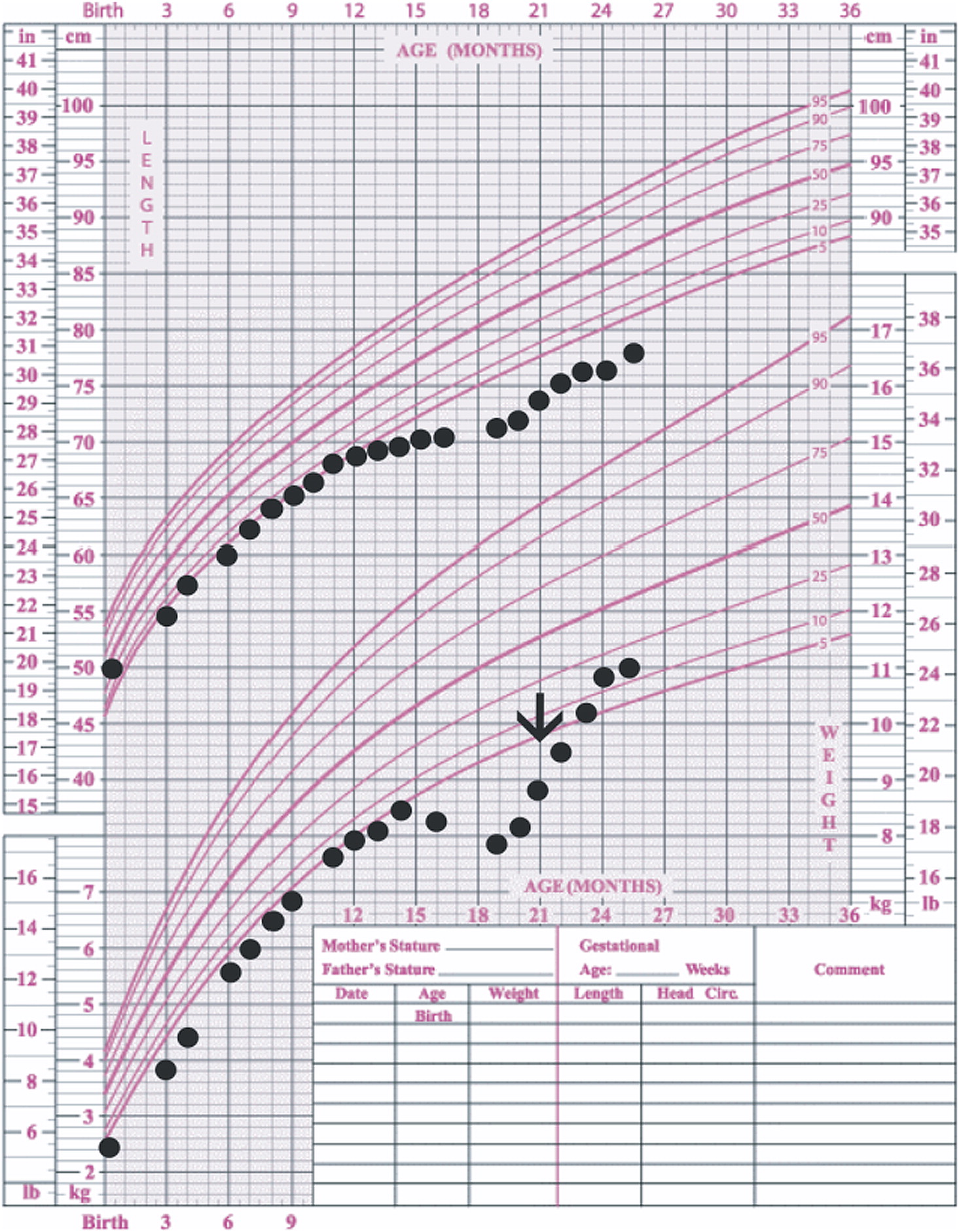
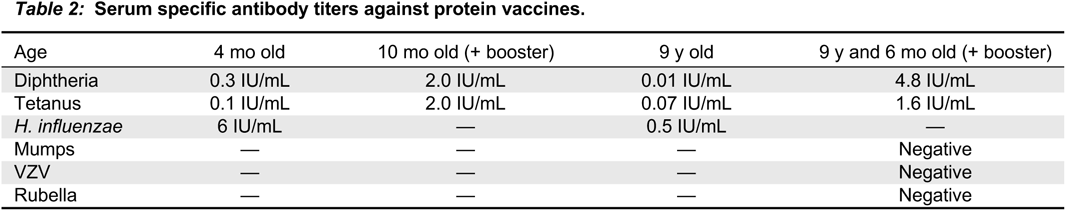
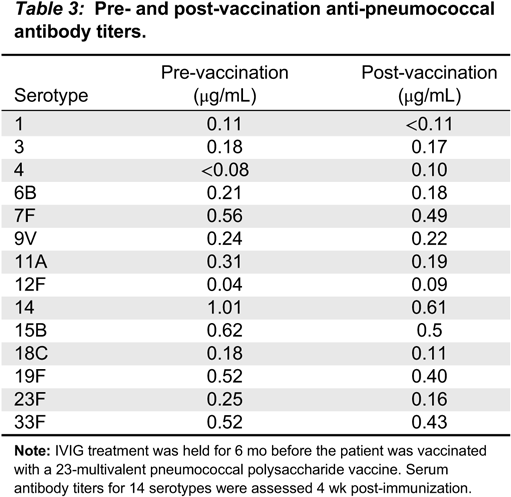
In our patient, both IgM and IgG serum levels were low and treatment with immunoglobulin was associated with reduction in the frequency of infections, elimination of septic episodes, and improved weight gain. She failed to develop a protective response to unconjugated pneumococcal capsule antigens despite adequate post-vaccine responses to tetanus and diphtheria, suggesting the diagnosis of specific antibody deficiency. The lack of specific polysaccharide antibodies has been previously reported in a patient with 18p- who only responded once immunized with a conjugated vaccine (
Browning 2010). Meanwhile, a recent report of a patient with ring chromosome 18 depicted a different vaccine response: while all immunoglobulin isotypes, including IgA, were low, he had a good responses to unconjugated pneumococcal vaccination and booster doses of diphtheria and tetanus vaccines (
Calvo Campoverde et al. 2016). Therefore, it is likely that, at least for protein vaccines, a booster dose can augment the specific antibody response in patients with chromosome 18 abnormalities and the use of conjugate vaccines should be prioritized.
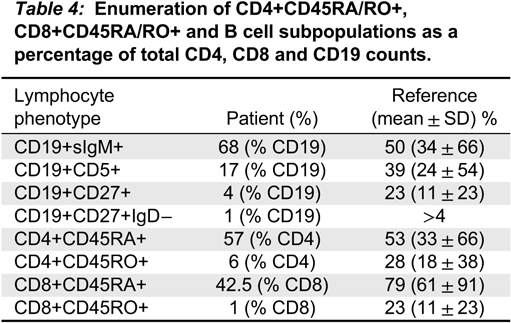
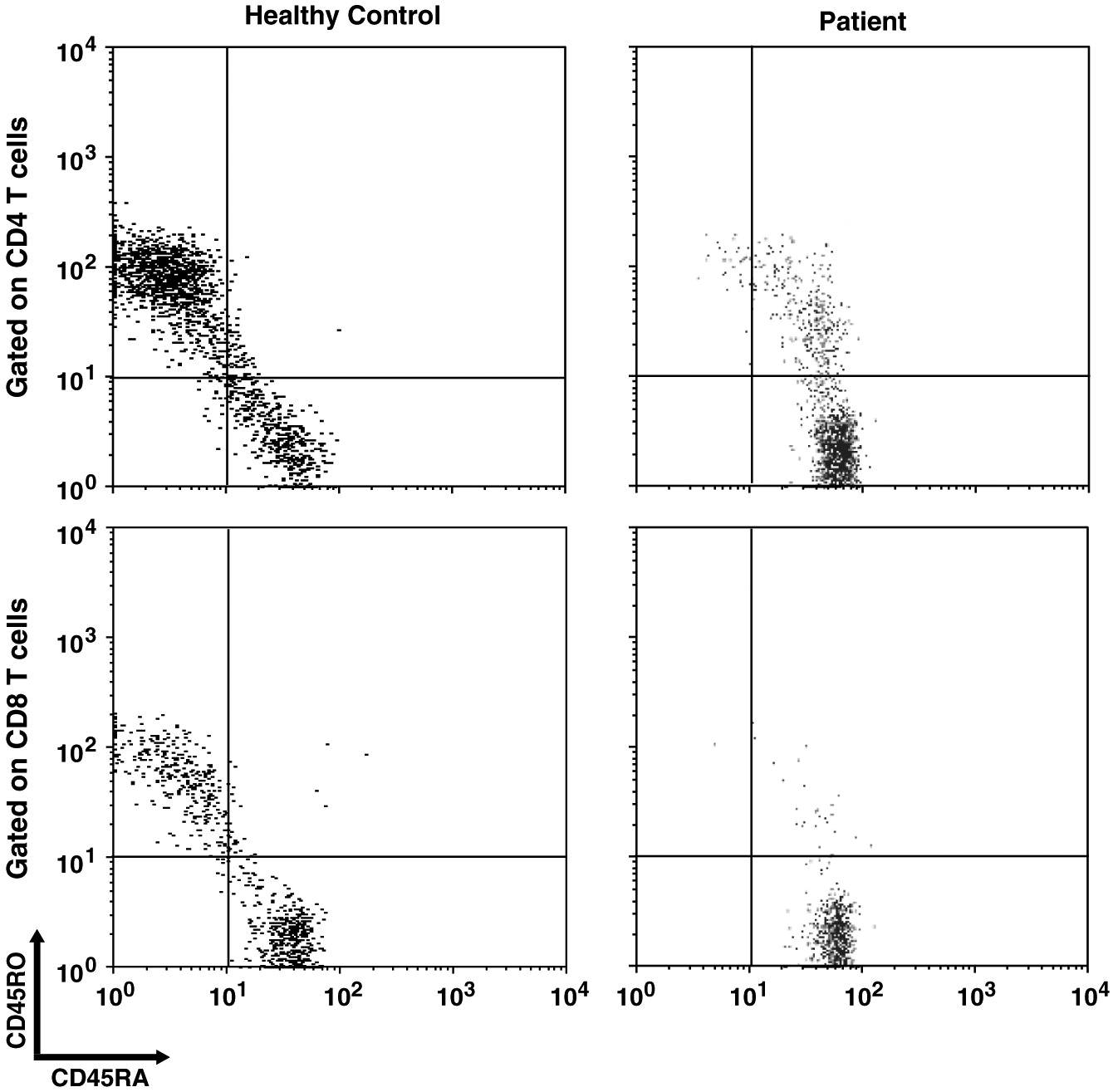
Impaired homeostasis of memory T and B cells has been documented in CVID patients with autoimmunity (
van de Ven and Warnatz 2015). We observed decreased CD19+CD27+ switched memory B cells and CD45RO+ memory T cell populations, as has been shown in 1 other patient with ring chromosome 18 (
Calvo Campoverde et al. 2016). CVID patients with reduced number of CD45RO+ T cells might be affected with severe viral infections (
Narula et al. 2007).
Association of primary antibody deficiencies and autoimmunity has been well characterized (
van de Ven and Warnatz 2015). In addition to humoral immunodeficiency, patients with chromosome 18 abnormalities have been affected with rheumatoid arthritis, lupus, thyroiditis, vitiligo and type I diabetes mellitus (
Dacou-Voutetakis et al. 1999;
Hasi-Zogaj et al. 2015). In a patient with a terminal deletion of 18q (18q21.32-q23) and low serum IgA and IgG4 levels, autoimmune thyroiditis and type 1 diabetes mellitus, regulatory T cell (Treg) counts were low (
Hogendorf et al. 2016). Our patient has severe polyarthritis, uveitis, and hypothyroidism. Unfortunately, we do not have data on Tregs in our patient.
The lack of efficient B cell and T cell interplay has been proposed to affect the development of memory T cells (
Martini et al. 2011). Our understanding of the genetics of B cell deficiencies over the past decade has largely advanced. None of those genes is located on chromosome 18. In fact, patients with proximal interstitial deletions of 18q do not show any abnormal immune phenotype (
Kato et al. 2010;
Imataka et al. 2015). Therefore, it is probable that the candidate genes for antibody defects on 18q are located distally. In fact, homozygous pathogenic variants of Mucosa-Associated Lymphoid Tissue Lymphoma Translocation 1 gene (
MALT1), that resides on 18q21.32, has been reported in patients with both CVID phenotypes with reduced switched memory B cells and autoimmunity with decreased Foxp3+ T cells (
McKinnon et al. 2014;
Charbit-Henrion et al. 2017). To examine if our patient’s immune dysregulation was due to a gene variant on the regions of deletion, including
MALT1, and to exclude other known monogenic causes of immunodeficiency and autoimmunity, we performed WGS. However, we could not find any variant that could explain the immunological phenotype, nor could we find any pathogenic variant, including CNVs, of
MALT1. To our knowledge,
MALT1 haploinsufficiency has not been reported in humans as yet.
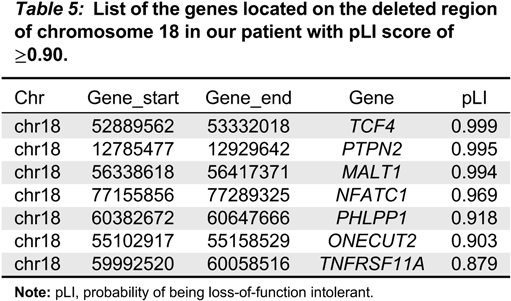
Indeed, few genes on chromosome 18 have been predicted to be haploinsufficient (
Cody et al. 2009), among which only
TCF4 falls into the deleted region in our patient. Heterozygous mutations of
TCF4, however, are associated with Pitt-Hopkins syndrome with no well-established immunological presentation. We ranked the genes on the deleted regions, using the probability of being loss-of-function (LoF) intolerant (pLI), described by the Exome Aggregation Consortium (ExAC;
exac.broadinstitute.org) (
Lek et al. 2016). Genes with pLI ≥ 0.9 are considered as an extremely LoF intolerant, meaning haploinsufficient.
We reviewed published studies in
Pubmed.com and further shortlisted genes of interest based on their possible involvement in immunological processes (
Table 5). Interestingly, both
TCF4 and
MALT1 ranked on the top of the list (
Table 5). This observation suggests that the phenotypical consequence of genetic variants of those genes might not have been recognized, including possible digenic conditions.
We also examined other genes in the deleted region, determined from the literature, that could contribute to the lack of memory cells and SIgAD.
PTPN2 is a negative regulator of Jak1 and Jak3 and deficient lymphocytes show increased STAT1 and STAT5 signaling (
Simoncic et al. 2002).
Ptpn2−/− mice show impaired B cell lymphopoiesis, as well as impaired T and B cell response to mitogens. However, T cell development in the thymus is not affected (
You-Ten et al. 1997). On the other hand, PTPN2 expression is enhanced in B cell lymphomas (
Lu et al. 2007). Importantly, it has been shown that early maturation of B cells in the bone marrow of
Ptnp2−/− mice was blocked due to enhanced IFNγ-STAT1 signaling (
Bourdeau et al. 2007). STAT1 activation is known to be upregulated in autoimmune disorders (
Domeier et al. 2016). Also, gain-of-function (GOF) mutations of
STAT1 have been shown in patients with CVID and autoimmunity (
Al Rushood et al. 2013;
Toubiana et al. 2016). Given the direct effect of PTPN2 on STAT1 phosphorylation (
ten Hoeve et al. 2002), loss of PTPN2 might contribute to a similar phenotype as of STAT1 GOF mutations.
TNFRSF11A, also known as Receptor Activator of Nuclear Factor Kappa-B (RANK), encodes the receptor for RANK ligand (RANKL) that together make the master signaling pathway for osteoclast differentiation. Patients with Autosomal-Recessive Osteoporosis (ARO) are affected by homozygous mutations of
RANK and
RANKL. Additionally, these patients are unable to produce antibodies in response to tetanus vaccination (
Guerrini et al. 2008). Interestingly, patients with homozygous mutations of
TNFRSF11A (
RANK), but not
TNFSF11 (
RANKL) show decreased switched memory B cells (IgD−CD27+), but did not show T cell abnormalities (
Guerrini et al. 2008).
PHLPP1 is a phosphatase that in parallel with PTEN regulates the PI3K/AKT pathway (
Chen et al. 2016). Heterozygous mutations of
PTEN cause Hamartoma Tumor Syndromes (PHTS). Patients with PHTS have defective antibody responses and autoimmune manifestations (
Driessen et al. 2016). While absolute numbers of transitional peripheral B cells are elevated in these patients, the CD27+ memory B cell population is decreased. Moreover, class switch recombination and somatic hypermutation are impaired in PHTS patients (
Driessen et al. 2016) as well as in PTEN deficient mice (
Suzuki et al. 2003). The effect of loss of PTEN on B cells has been attributed to increased PI3K/AKT signaling. In fact, in patients with dominant GOF mutations of P110δ (a subunit of PI3K electively expressed in lymphocytes) show impaired class switch recombination, defective antibody responses and increased transitional B cells (
Angulo et al. 2013). Notably, although loss of PTEN in humans does not affect Treg development, inhibitors of PHLPP1 block in vitro Treg differentiation (
Chen et al. 2016). Collectively, absence of PHLPP1 in patients with 18q- could putatively replicate the immunological phenotype of PTEN deficiency in association with defective Treg responses. We found a deep intronic 4 kb deletion in intron 8 of
PHLPP1 in our patient. However, this deletion is very unlikely to be deleterious as it is quite far from flanking splice sites. Further functional studies will be required to assess whether haploinsufficiency of
PHLPP1 is could lead to this immunological phenotype.
NFATC1, encoding the nuclear factor of activated T cell c1 (NFATc1), is a member of NFAT family of transcriptional factors that are regulated via Ca
2+/Calcineurin (
Medyouf and Ghysdael 2008). Based on animal models, it has been suggested that NFATc1 deficiency might cause immunodeficiency in human patients with 18q- (
Li et al. 1995). Loss of
Nfatc1 in mice affects development and survival of both peritoneal and splenic B1-a cells (
Berland and Wortis 2003). Moreover, even though NFATc1 does not affect the development and maturation of B cells, it plays a critical role in their function and fate. It has been shown that
Nfact1−/− B cells in mice show impaired proliferation and survival in response to BCR stimulation, Ig class switching and can suppress T cell activation (
Bhattacharyya et al. 2011). These findings were in association with impaired Ca
2+ influx resembling defective BCR signaling (
Bhattacharyya et al. 2011). On the other hand, loss of NFATc1 in T cells impaired homing of follicular regulatory T cells in B cell follicles via transactivation of CXCR5. This was associated with an exacerbated lupus-like phenotype in mice (
Bhattacharyya et al. 2011).