Introduction
The Forkhead-box (FOX) superfamily of transcription factors play important roles in the homeostasis, function, and aging of a variety of organs and tissues – including the immune system. FOX protein N1 (FOXN1), encoded by the
FOXN1 gene, is an essential regulator of thymic epithelial cell differentiation required for T cell development, and is also expressed in skin and hair follicles (
Vaidya et al. 2016). Potential roles in hematopoiesis within the bone marrow have been reported (
Zook et al. 2013).
The thymus provides a unique environment for the differentiation, selection, and maturation of bone marrow-derived T cell progenitors into fully functional T cells. This process relies on the ordered architecture of the thymus, consisting of a cortical area rich in cortical thymic epithelial cells (cTECs) and lymphoid cells, a medullary area containing medullary thymic epithelial cells (mTECs), Hassall’s corpuscles, macrophages, dendritic cells and B lymphocytes, as well as a transitional corticomedullary junction. Importantly, activation of FOXN1 and subsequent transcriptional regulation of downstream genes are needed for the proper differentiation, maturation, and function of cTECs and mTECs during prenatal thymus organogenesis, as well as homeostasis of the postnatal thymus (
Corbeaux et al. 2010;
Vaidya et al. 2016). Thus, loss-of-function of this so-called master regulator results in ablated thymic and T cell development, and the phenotype of severe combined immunodeficiency (SCID).
Bi-directional cues between the thymic stroma and T cells are essential for the normal development and function of both entities (
van Ewijk et al. 1994). Defective thymic architecture has been documented in instances where T cells exhibit stunted development (
Palmer et al. 1993), whereas reconstitution of the thymic microenvironment has been reported with the administration of T cell-committed precursors (
Roberts et al. 2009). Importantly, animal models have been an essential tool for dissecting the role of FOXN1 during early thymus development (
Bosticardo et al. 2019). Mice deficient in
Foxn1 (‘nude’ spontaneous mutation) are athymic, lack T cells, and exhibit abnormal hair growth (
Nehls et al. 1994). Among the panel of genes regulated by foxn1 are
Ccl25, required for homing of immature thymocytes to the thymus (
Liu and Leung, 2006),
Dll4, involved in the Notch pathway-directed sorting of cells towards T cell lineage (
Koch et al. 2008), and
Cxcl12, needed for expansion of T cell precursors in the thymus (
Ara et al. 2003). Other genes induced by foxn1 required for thymic epithelial cell and thymocyte exchange are
Psmb11,
Prss16, and
Cd83 (
Bosticardo et al. 2019).
In humans, null mutations in
FOXN1 are characterized by severe T cell immunodeficiency (T-B+NK+ phenotype), congenital alopecia, and nail dystrophy (OMIM #601705) (
Amorosi et al. 2008;
Frank et al. 1999;
Pignata et al. 1996). Affected individuals suffer recurrent infections, failure to thrive, and oral candidiasis. While some attempts to correct the deficiency have been made with hematopoietic stem cell transplantation (HSCT), overall, post-transplant outcomes for individuals with defects in
FOXN1 indicate that thymus transplants are a more effective and curative option (
Bosticardo et al. 2019;
Chou et al. 2014).
Monoallelic mutations in
FOXN1 are more frequently associated with variable clinical features. A recent longitudinal analysis of 25 children and 22 adults with heterozygous
FOXN1 variants described the majority being clinically well except for viral respiratory infections (
Bosticardo et al. 2019). Common non-infectious manifestations included eczema and nail dystrophy. Of the children, 21 were identified following an abnormal newborn screen (NBS) result and were associated with T cell lymphopenia. For most of these cases, CD4+ T cells eventually recovered.
Here, we report on our experience with an infant who was diagnosed with T-B+NK+ severe combined immunodeficiency (SCID) following an abnormal NBS result. While undergoing work up for HSCT, extensive genetic investigations including whole exome sequencing (WES) and whole genome sequencing (WGS) identified a novel heterozygous variant in FOXN1, which subsequently changed the management plan for this patient.
Case presentation
The patient is currently a 19-month-old female, the first child to non-consanguineous parents of Asian descent. The pregnancy was uncomplicated and she was born at 39+2 weeks gestation by spontaneous vaginal delivery. Her mother had 2 prior early miscarriages and her father has 2 healthy older children from a previous marriage. The patient came to the attention of the Immunology team following an abnormal NBS result for SCID. The initial T cell receptor excision circle (TREC) level was 0 (cut-off: 75), and the repeat TREC was 7. Immunodeficiencies associated with ADA, PNP, ZAP70, and IKBKB were ruled out during initial screening tests, and assessment for microdeletions in 22q11.6 was returned negative.
Initial Evaluations
Initial immune evaluation revealed T cell lymphopenia of both CD4+ cells (43 cells/μL, normal: 2330-3617 cells/μL) and CD8+ cells (5 cells/μL, normal: 712-1361 cells/μL). B cells (431 cells/μL, normal: 315-1383 cells/μL) and NK cells (622 cells/μL, normal: 201-870 cells/μL) were within normal range. Output of naïve T cells was low at 9% (CD4+/CD45RA+), while memory T cells predominated at 84% (CD4+/CD45RO+). Recent thymic emigrants were low at 9% (normal: 64–94%). Her lymphocyte proliferation responses to the mitogen phytohemagglutinin (PHA) were very low at 12 (stimulation index, SI normal >400) (
Table 1). Ultrasound of the neck revealed normal size lymph nodes in both jugular chains; however, there was no convincing evidence of thymic tissue.
Genetics
Genetic work-up to identify a causative molecular defect underlying the patient’s immunodeficiency was started. An initial SCID gene panel (Newborn Screening Ontario; 20 genes) was negative. Subsequently, a more extensive PID panel (Blueprint Genetics) was sent which identified, at 2 months of age, a novel heterozygous frameshift mutation in FOXN1 (NM_003593.2), c.1465del, resulting in p. Gln489Argfs*61. This variant was absent from large population databases (gnomAD). To ensure that no other genetic aberrations were present, trio WGS of the patient and her parents were sent. While no further changes were found (including non-coding or copy number variants), the mutation was identified as a de novo variant targeting exon 7 — a region where many pathogenic variants reside.
Management
Our patient’s initial presentation was in keeping with T-B+NK+ SCID, and she was managed accordingly with antibiotic prophylaxis and immunoglobulin replacement therapy (from 4 weeks of age) while workup for HSCT was initiated. After the pathogenic mutation in FOXN1 was identified, in line with our center’s experience with patients harboring heterozygous variants in FOXN1, we instead changed our approach to ‘watchful waiting’ to see if her immune parameters would improve.
In the subsequent months, the patient’s lymphocyte function recovered (PHA stimulation index increased gradually to 481.7), and her T cell counts continue to show improvement although are still low (
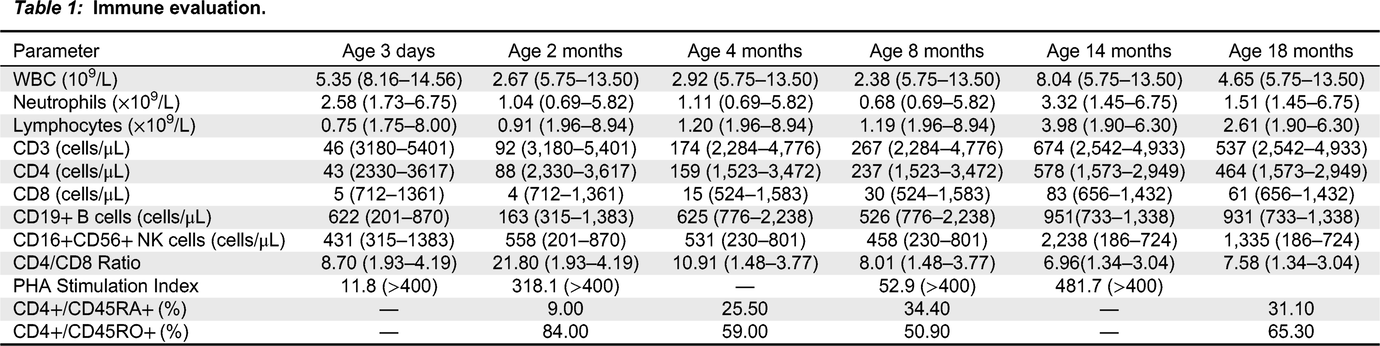
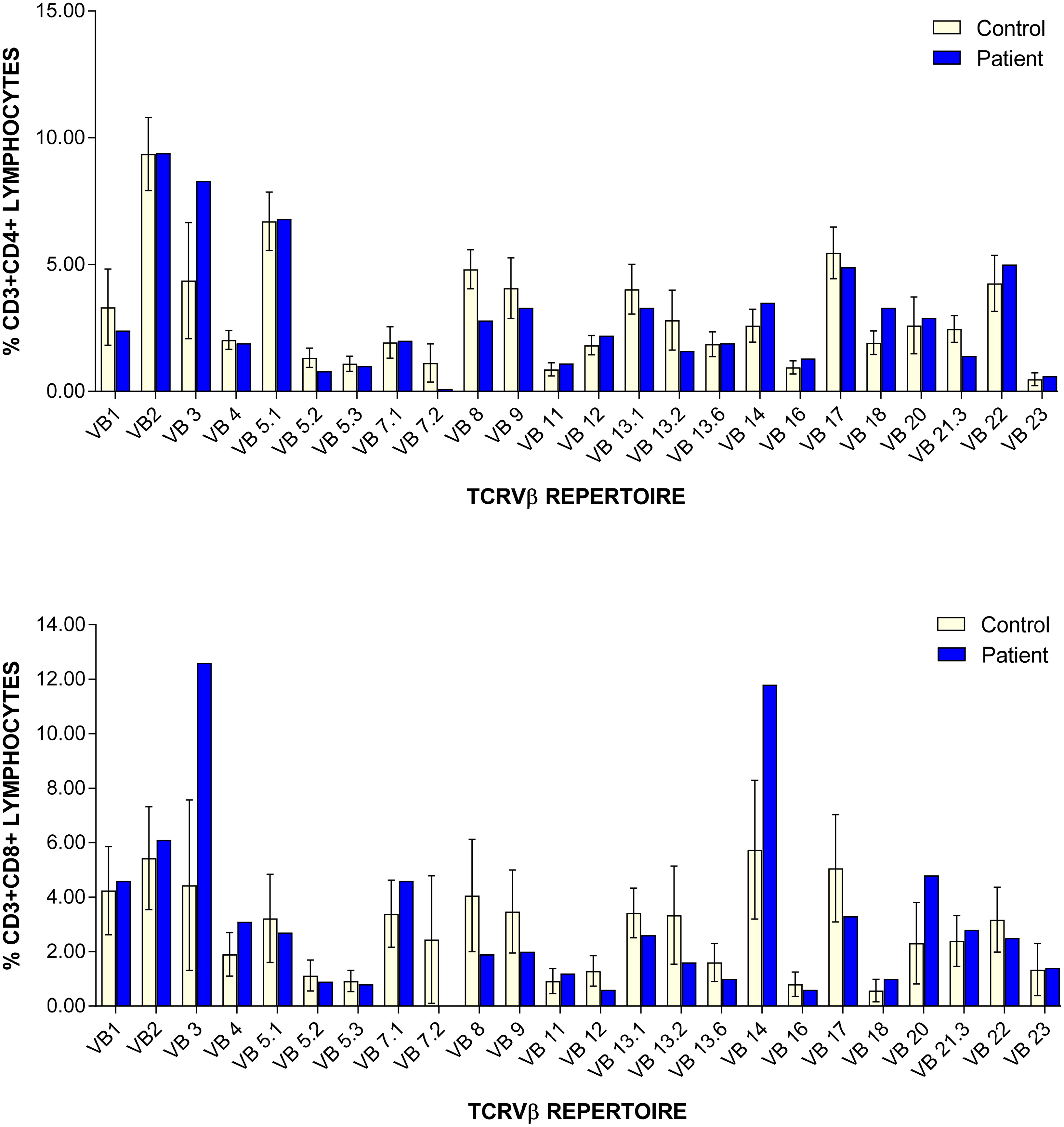
Table 1). CD45RA/RO assessment revealed improving naïve T cells (31.10%) versus memory T cells (65.30%). Recent thymic emigrants are steadily increasing 21% (normal: 40–100%). Assessment of TCRVβ repertoire revealed only slight under-representation of CD4+ Vβ8 and Vβ21.3 clones, and over-representation of CD8+ Vβ3 and Vβ14 clones (
Figure 1). At 8 months of age, antibody replacement therapy was stopped and her immunoglobulin levels are currently within normal range (IgG: 5.1 g/L (normal: 3.2–11.5 g/L); IgA: 0.2 g/L (normal: 0–0.9 g/L); IgM: 0.5 g/L (normal: 0.5–1.9 g/L)). She has received her routine non-live vaccines and is responding well (anti-tetanus toxoid IgG >5 IU/mL).
To date, the patient remains clinically well and has not had any significant infections. There is presently no indication for HSCT and she continues to be followed closely.
Discussion
The implementation of SCID NBS has transformed our ability to identify and treat infants with SCID before infectious complications and end-organ damage occurs (
Kwan et al. 2014). Screening utilizes measurement of TRECs, which are a by-product of T cell receptor recombination. Thus, very low/absent TRECs in neonates are an indication of defects in T cell development and prompts the diagnostic evaluation for SCID (
Biggs et al. 2017). By extension, less profound immunodeficiencies that would not have been detected by past practices until individuals presented clinically are also being flagged for follow-up (
Amatuni et al. 2019;
Mandola et al. 2019;
Scott et al. 2021).
Our patient was initially diagnosed with T-B+NK+ SCID following abnormal SCID NBS with two absent/low TREC determinations. While she underwent workup for HSCT, extensive genetic investigations identified a single novel de novo heterozygous mutation in
FOXN1. Our center’s experience with heterozygous
FOXN1 mutations (
Scott et al. 2021) as well as those reported in the literature, changed the intended management plan to a more conservative stance given the likelihood that the patient’s immune parameters would improve with age. Moreover, HSCT is unlikely to correct the lymphopenia associated with a poorly developed thymus (
Bosticardo et al. 2019;
Chou et al. 2014). Our case is unique in that the genetic work-up involved gene panel sequencing, WES, and WGS to rule out the possibility of another underlying genetic etiology, affirming our approach to closely monitor the patient for improvement.
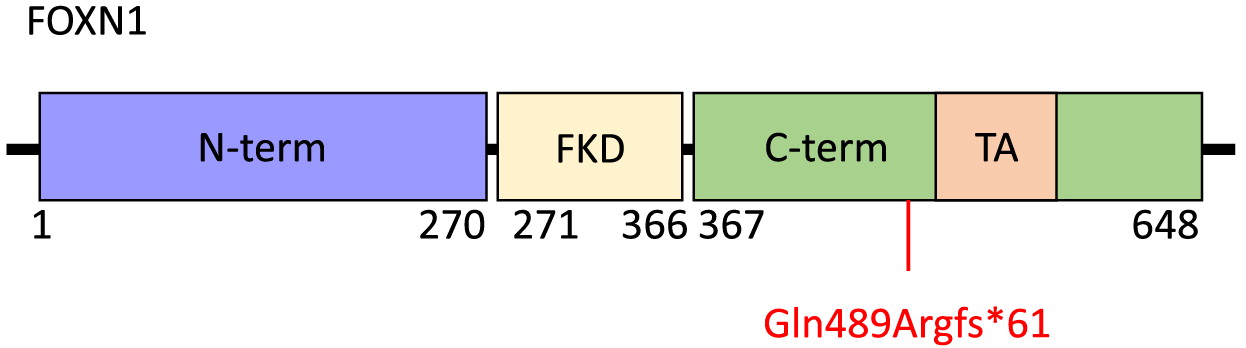
FOXN1 is a 648-amino acid protein with an N-terminal region of residues 1–270, a Forkhead domain located centrally between amino acids 271 and 366, and a C-terminal region comprising residues 367–648 (
Figure 2). Within the C-terminal region is an evolutionarily conserved transcriptional activation domain. The N-terminal region has been implicated in thymic epithelial cell differentiation while the C-terminal region is required for high-affinity DNA binding (
Newman et al. 2020). The
FOXN1 frameshift mutation identified in our patient, c.1465del (p.Gln489Argfs*61), targets exon 7 and causes a premature stop at position 61 within the C-terminal domain. This is predicted to result in loss of normal protein function due to truncation of the protein or nonsense-mediated mRNA decay. The clinical picture of our patient, with CD4+ and CD8+ T cell lymphopenia and unaffected B and NK cell counts is not dissimilar to other reports of patients with mutations within the C-terminal domain, nor of other domains within FOXN1 (
Bosticardo et al. 2019). Thus, it remains to be seen whether a genotype-phenotype correlation exists.
Athymia has been reported in individuals with pathogenic homozygous
FOXN1 mutations (
Chen et al. 2009) as well as heterozygous variants (
Bosticardo et al. 2019). Of the 13 infants reported by Bosticardo and colleagues, only 3 had a normal-sized thymic shadow while the remaining were absent or reduced in size. In our patient, ultrasound did not detect evidence of thymic tissue. Nevertheless, the presence of near normal TCRV beta repertoire indicates that the thymus is likely small or perhaps ectopically placed (
Shah et al. 2001).
In summary, this report highlights the importance of robust genetic investigations to delineate the underlying cause of immunodeficiencies detected by SCID NBS. We continue to follow a conservative approach with this patient, particularly as more reports of disease course and outcomes of individuals affected by heterozygous FOXN1 mutations come to light.