Background
The most common chromosomal microdeletion disorder identified in children is 22q11.2 deletion syndrome (22q11.2DS), whereby affected individuals are missing a small piece of chromosome 22 containing genes responsible for midline fusion during the embryological period. The syndrome results in a constellation of signs and symptoms, described as DiGeorge or Velocardiofacial Syndrome. Some of the clinical features include congenital heart disease, cleft palate, hypoparathyroidism, thymic hypoplasia, immunologic abnormalities, developmental delay, psychiatric disorders, and distinct facial features (
McDonald-McGinn et al. 2015).
The most common deletion responsible for the syndrome, present in 85% of individuals, involves 3 megabases (Mb) in the 22q11.2 chromosomal region (
McDonald-McGinn et al. 2013). This region contains 90 known or predicted genes, including the T-box transcription factor (
TBX1) gene. The
TBX1 gene is implicated in several of the clinical features of the syndrome. Most of the remaining patients have smaller 1.5 Mb deletions, referred to as atypical “nested” deletions (
McDonald-McGinn et al. 2015).
Suggestive clinical presentations have traditionally prompted evaluation for 22q11.2DS through the use of fluorescent
in situ hybridization (FISH) using specific probes, including TUPLE or N25. Chromosomal microarray (CMA) and multiplex ligation-dependent probe amplification (MLPA) can also be used, particularly to identify smaller nested deletions not detectable by FISH (
McDonald-McGinn et al. 2015).
To allow for earlier detection and diagnosis, the inclusion of 22q11.2DS in newborn screening (NBS) panels has been proposed (
Pretto et al. 2015). However, routine screening has not yet been implemented. Given that immune dysfunction, specifically low or absent T-cells, is present in some cases of 22q11.2DS, some patients can be identified by NBS for severe combined immune deficiency (SCID), through detection of low T-cell receptor excision circles (TRECs). TRECs are a biomarker of naïve T-cells and their absence signifies low or absent T-cell production (
Kwan and Puck 2015). The TREC screen primarily aims to identify forms of SCID; however, it also identifies infants with non-SCID T-cell lymphopenia. The differential for these secondary targets is broad, and includes syndromes such as 22q11.2DS, other combined immune deficiencies, as well as cardiac and gastrointestinal diseases and lymphatic malformations (
Dorsey and Puck 2017).
In the last 4 years, SCID NBS has been implemented across North America, in Ontario, the Canadian Maritime provinces, and most American states. In Ontario, term infants (defined as ≥33 weeks gestational age and ≥1500 g) with a TREC level falling below a set cutoff, falling significantly below the normal range, will screen positive for SCID. Further screening, including a purine profile and a quantitative polymerase chain reaction (q-PCR)-based TBX1 deletion analysis to screen for 22q11.2DS, is subsequently performed on the NBS samples from this group of infants. Infants with a screen positive result for SCID are referred for additional investigation. Follow-up testing typically includes a repeat dried blood sample for repeat TREC, TBX1, and purine profile screening. Diagnostic follow-up often includes a complete blood count with a differential, lymphocyte immunophenotyping, testing for 22q11.2DS via FISH or CMA, and consultation with a medical expert. Follow-up testing practices vary across centres.
Herein, we report a case of a newborn with 22q11.2DS detected by low TRECs on the SCID NBS, with a normal screening TBX1 result. A deletion involving the TBX1 gene was subsequently detected on our centre’s standard follow-up 22q11.2 FISH, and by subsequent CMA. This case highlights the limitations of the TBX1 screening assay and the importance of performing diagnostic testing with FISH and (or) CMA regardless of the initial TBX1 result.
Case presentation
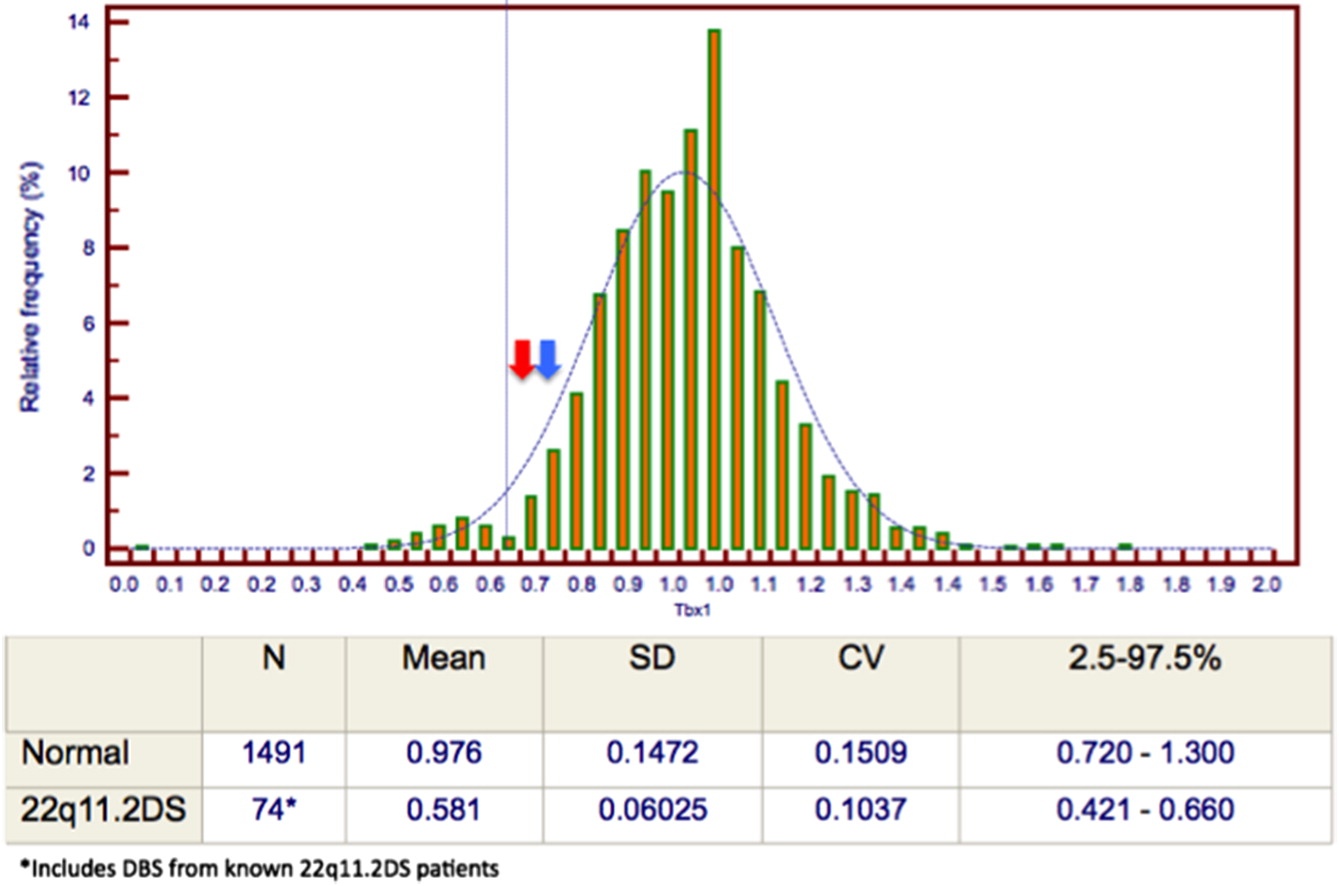
A newborn male born after an uncomplicated, non-consanguineous pregnancy was born at 38 weeks gestation with a birth weight of 2390 g. NBS done at 24 hours of life was positive for SCID. Initial screening revealed a low TREC value of 54 copies/3 μL (cutoff: 75, reference interval: 228–2634), normal purine profile and a normal TBX1 analysis with no deletion detected. Upon assessment in our Pediatric Immunology Clinic, he appeared well. Physical examination demonstrated a non-dysmorphic infant with intact palate, palpable lymph nodes, and cardiovascular examination within normal limits. The remainder of the examination was unremarkable. The TREC screening assay was repeated and the value remained low at 51 copies/3 μL on the follow up sample, with a normal purine profile and TBX1 analysis. Additional investigations included a complete blood count with differential and lymphocyte immunophenotyping, which were within normal range.
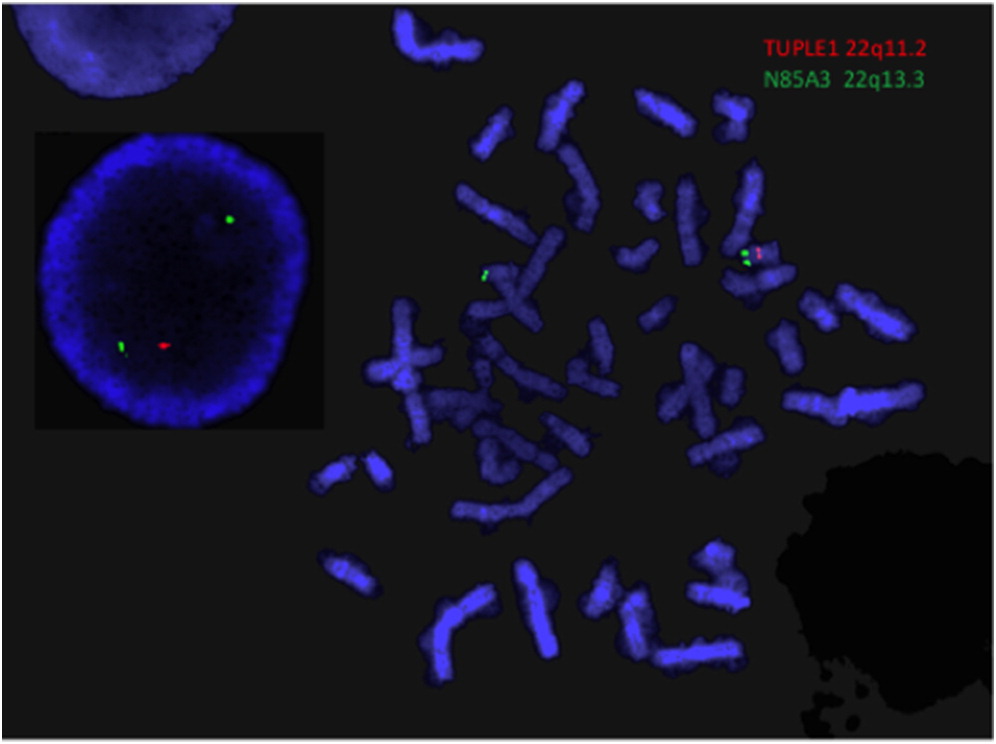
Despite the normal
TBX1 screening analysis, a FISH assay to assess for 22q11.2DS was performed as part of the subsequent routine testing protocol at McMaster Children’s Hospital. A deletion in the 22q11.2 region was identified on FISH (
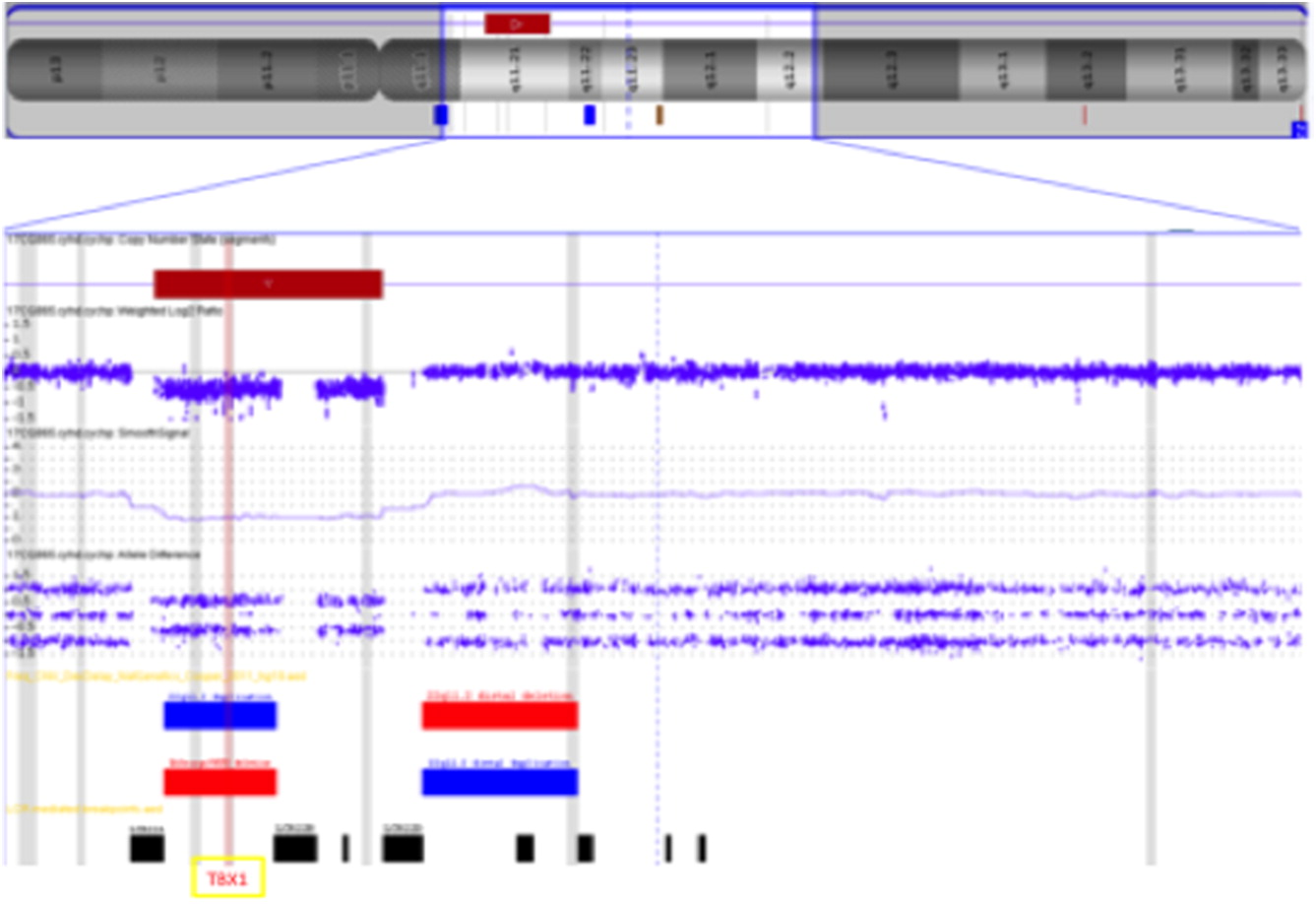
Figure 1). A CMA was subsequently performed to further delineate the deletion, which detected a 2.5 Mb microdeletion within the typical 3 Mb region, including the
TBX1 gene. This confirmed the diagnosis of 22q11.2DS (
Figure 2) for this patient. An echocardiogram was performed, revealing an aberrant right subclavian artery and vascular ring. Serum calcium and parathyroid hormone levels were within normal range. No abnormalities were noted on renal ultrasound. The baby remains clinically well and will have ongoing follow-up in the Immunology and 22q clinics at McMaster Children’s Hospital.
Conclusion
This case of 22q11.2DS with a typical deletion encompassing the TBX1 gene not identified by initial TBX1 screening highlights the limitations of this test as a standalone screening assay. To our knowledge, this is the first such case to be described in the literature. As the diagnosis of 22q11.2DS has broad implications for further investigation, management and counselling, we propose that diagnostic testing with FISH and (or) CMA for 22q11.2DS is required after a NBS demonstrating low TRECs, regardless of the result of the TBX1 screening analysis.
As routine screening for SCID in Canada only began in 2013, data regarding process and outcomes remain limited. The described case demonstrates a need for a standardized follow-up testing algorithm across Canadian institutions for newborns who screen positive for SCID, which should include definitive diagnostic testing for 22q11.2DS. Further study will help to determine a comprehensive protocol to follow a low TREC result.