A novel mutation in LIG4 in an infant presenting with severe combined immunodeficiency with thymic medullary dysplasia
Abstract
Introduction
Methods
Lymphocyte markers and T cell proliferative responses
Serum concentration of immunoglobulins
Whole exome sequencing and variant calling
Variant prioritization
Pathology
Results
Case report
Genetic evaluation
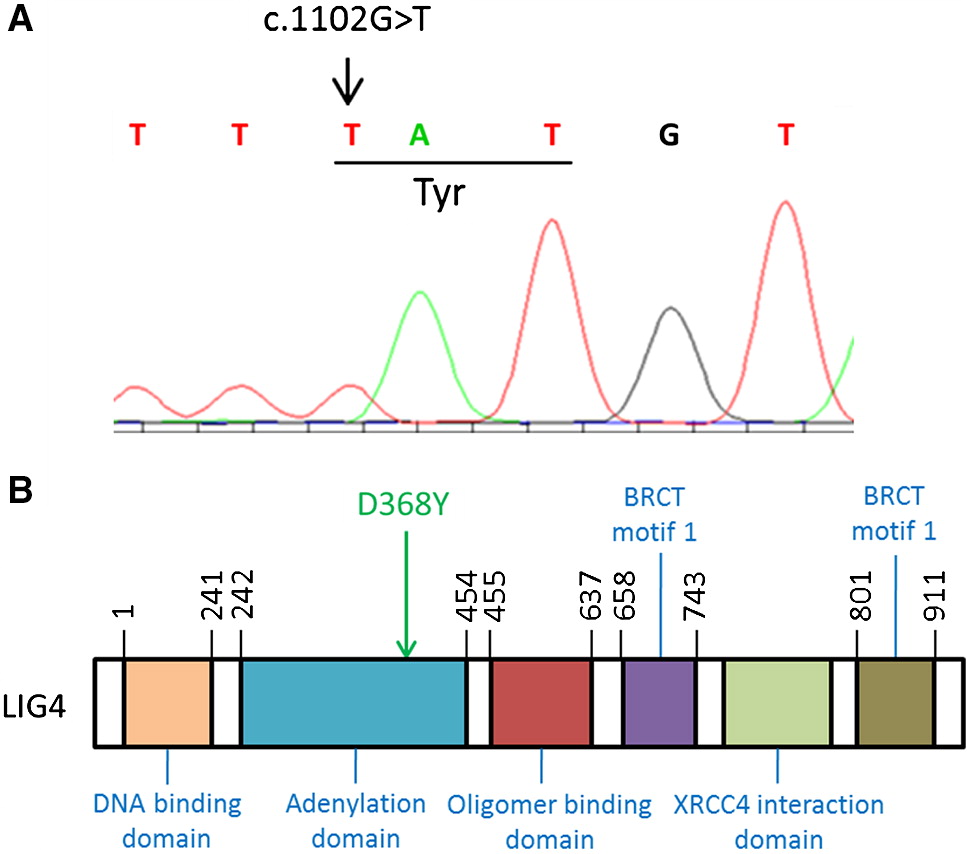
Immune evaluation
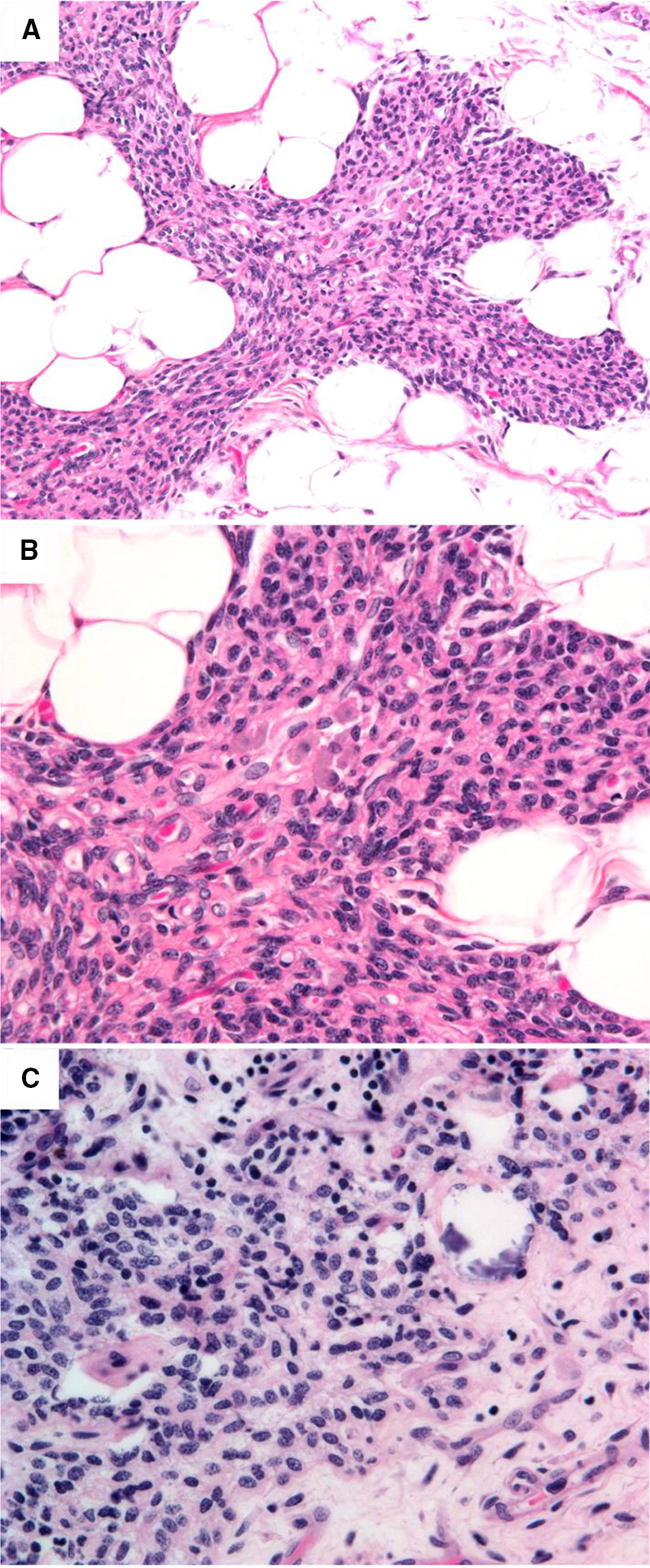
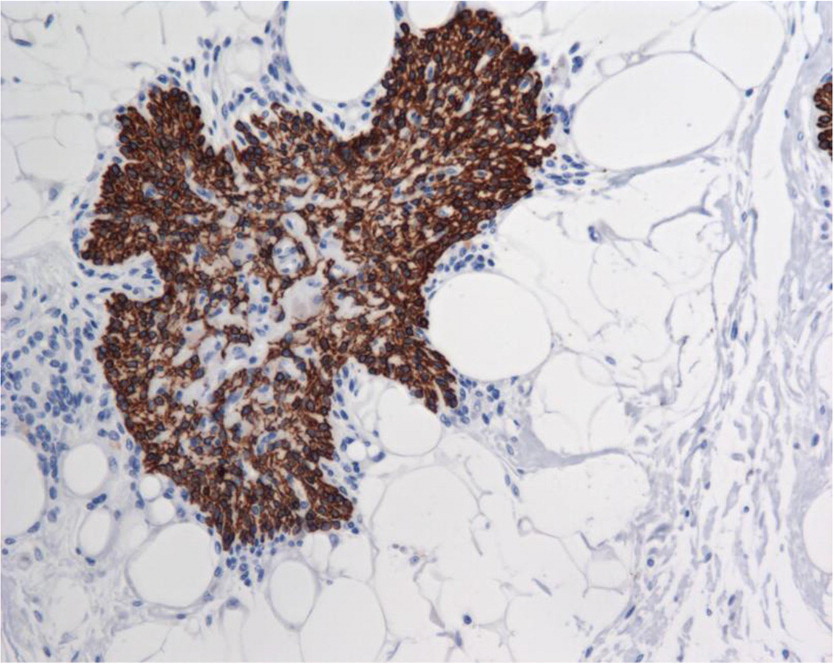
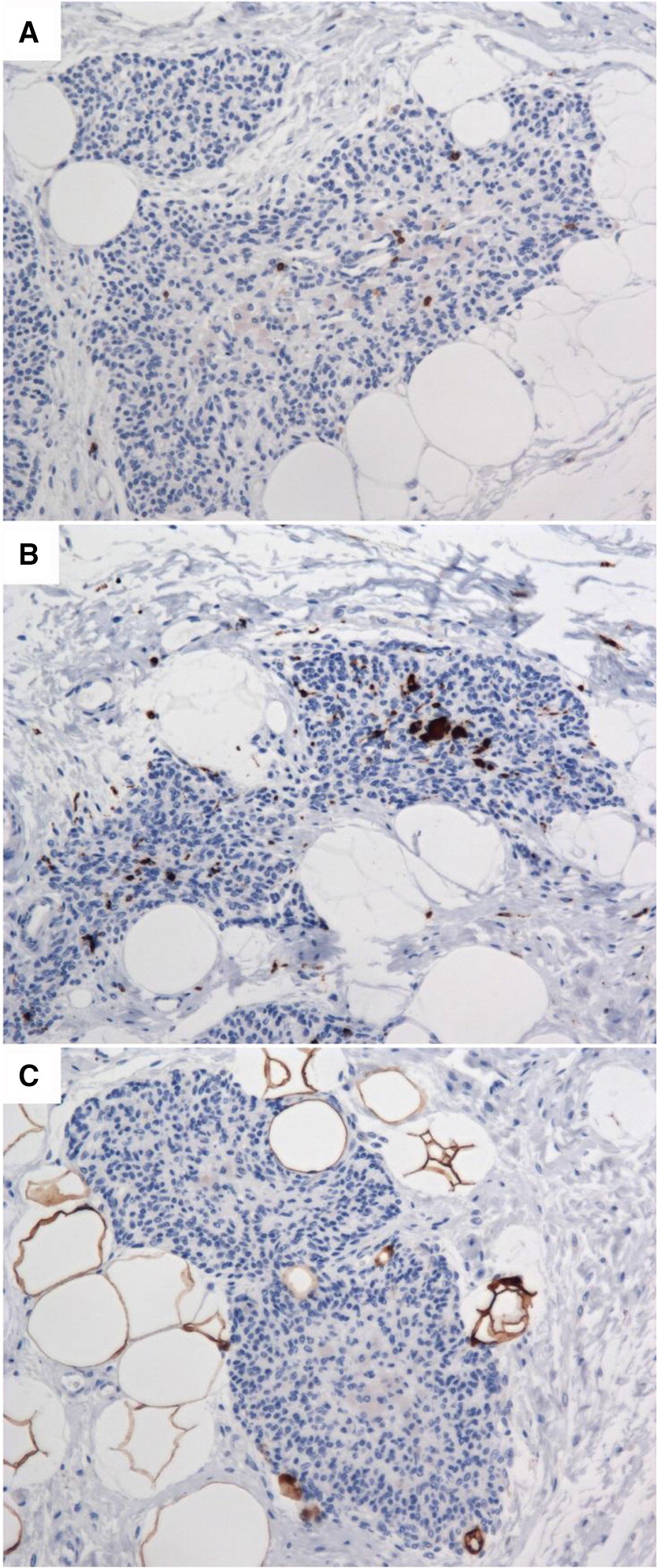
Thymus pathology
Discussion
REFERENCES
Information & Authors
Information
Published In

History
Copyright
Authors
Metrics & Citations
Metrics
Other Metrics
Citations
Cite As
Export Citations
If you have the appropriate software installed, you can download article citation data to the citation manager of your choice. Simply select your manager software from the list below and click Download.
There are no citations for this item
View Options
View options
Login options
Check if you access through your login credentials or your institution to get full access on this article.


