Introduction
The nuclear factor kappa-B (NFkB) family of transcription factors consists of 5 members: RelA (p65), RelB, Rel (c-Rel), and 2 precursor proteins NFkB1 (p105) and NFkB2 (p100), which are processed to p50 and p52, respectively (
Sun 2011;
Millet et al. 2013). Two major pathways of NFkB activation are recognized: the classical, mediated by RelA (and c-Rel); and the less understood alternative, mediated by RelB. Infectious agents (through toll receptors), inflammatory cytokines, and lymphocyte antigen receptor activation all activate NFkB activity (
Sun 2011;
Millet et al. 2013).
All the NFkB proteins contain a Rel homology domain necessary for DNA binding. However, trans-activation and nuclear localization domains are separated between proteins, requiring the formation of dimers to obtain an active complex with nuclear targeting ability. Classical pathway activation is regulated by the binding of inhibitory IκB proteins to Rel-A/p105 complexes, sequestering them in the cytoplasm (
Sun 2011;
Millet et al. 2013). Phosphorylation of IκB proteins by IKK kinases initiates IκB degradation, freeing the NFkB complexes to then enter the nucleus. In contrast, RelB activity is suppressed by the binding of NFκB2 p100; alternative pathway activation requiring inducible proteolytic processing of p100 to its p52 form (initiated by NIK and IKKα) to permit RelB nuclear entry (
Sun 2011;
Millet et al. 2013).
Although RelA is activated by a large number of receptors (e.g., TNFR, TLR4, TCR), RelB activity is observed downstream from only a handful (BAFF-R, CD40, TWEAK, Lymphotoxin-β-R), despite its wide expression. Furthermore, although these receptors utilize the alternative pathway, simultaneous activation of the classical pathway is observed. In addition to its receptor activated functions, the presence of RelB is proposed to be necessary for the regulation of other family members (
Oeckinghaus et al. 2011;
Sun 2011;
Millet et al. 2013).
RelB knockout mice show that the protein plays an important role in the development of secondary lymphoid tissue (
Burkly et al. 1995;
Weih et al. 1995). Furthermore, most
RelB −/− mice die young, owing to overwhelming multi-organ inflammatory processes stemming from aberrant T-cell cytokine secretion and (or) antigen recognition.
Methods
Patient data were compiled prospectively and retrospectively from medical records and were entered into the Canadian Centre for Primary Immunodeficiency Registry and tissue bank, which was approved by the SickKids Research Ethics board (protocol no. 1000005598). This includes consent and assent from patients and parents for genetic analysis and the collection of tissue including thymus. Patients were treated primarily at The Hospital for Sick Children, Toronto, Ontario.
Linkage analysis
MERLIN (
Abecasis et al. 2002) was used for parametric linkage analysis. Minor allele frequency was established based in HapMap CEU (North Americans and Northern and Western European descent) and TSI (Italians from Tuscany) samples. The parametric logarithm base 10 of odds (LOD) score was calculated for a 1 centiMorgan (cM) grid along the chromosomes at disease allele frequency of 1%.
Whole genome sequencing
The whole genome of 1 patient (patient 2) was sequenced using the Complete Genomics (Mountain View, California, USA) platform.
T- and B-cell proliferative responses
Lymphocyte proliferative responses to mitogens (including phytohemagglutinin (PHA) and anti-CD3 antibodies) and to a panel of recall antigens (including candida, tetanus, herpes zoster, and cytomegalovirus) were determined by thymidine incorporation at day 3 or day 6. All assays were performed in triplicate and were compared with simultaneously stimulated random normal controls.
Immunoglobulin and specific antibody determinations
Serum concentrations of immunoglobulins were measured by nephelometry. Serum IgE concentration was measured by radioimmunoassay with the IgE PRIST kit (Pharmacia Diagnostics, Quebec, Canada). Levels of serum antibodies to tetanus were measured by ELISA and polio antibody titers were determined by complement fixation.
Real-time polymerase chain reaction (RT-PCR)
RT-PCR was performed on an ABI 7500 real-time PCR system (Applied Biosystems) using 0.1µg of cDNA with the SYBR select Master Mix (Life Technologies). We used the following primers:
GAPDH-F: GAAACTGTGGCGTGATGGC,
GAPDH-R: CACCACTGACACGTTGGCAG,
RELB-F: CCCTACGAGGACCTGGAGAT, and
RELB-R: GCAGGAAGTGGTCCAGGAT.
Western blotting
Whole-cell lysates were prepared in a RIPA buffer and analyzed by Western blotting. Anti-RelA, c-Rel, IκBα, NFκB1 p100, actin, Giα, C-terminal RelB (sc-226), and N-terminal RelB antibody (sc-38007) were purchased from Santa Cruz Biotechnology Inc. (Dallas, Texas, USA). Anti N-terminal RelB (04-1077) was also purchased from EMD Millipore (Billerica, Massachusetts, USA). Where necessary, expression levels were quantified by densitometry scanning and normalized by comparison with matching control protein levels. All blots were repeated at least twice.
Results
Case reports
Patient 1 was born after cesarean section at 41 weeks to consanguineous parents of Irish descent. At the age of 4 months, patient 1 began experiencing persistent cough and had 3 episodes of pneumonia, all of which required hospitalization. In addition, he suffered multiple episodes of otitis media and required tympanostomy tubes.
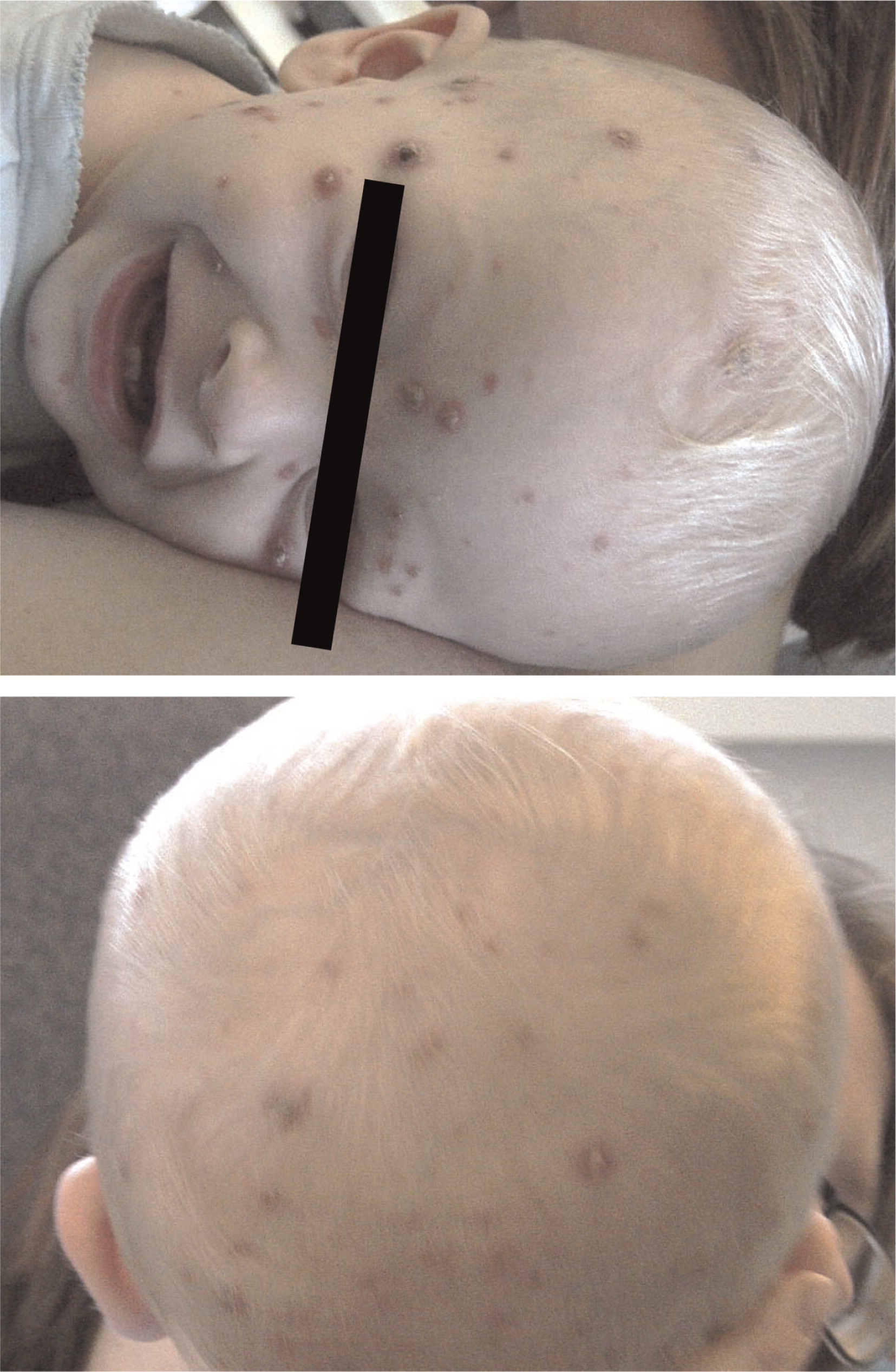
Patient 2, the younger brother of patient 1, was born at term by scheduled cesarean section. From the age of 1 month he had a chronic cough and repeated upper respiratory infections. At the age of 13 months, he was admitted for ecthyma gangrenosum which was confirmed by skin biopsy (
Figure 1). He has suffered multiple episodes of lobar pneumonia that required antibiotic treatment and hospitalization, as well as failure to thrive. He was noticed to have intermittent swelling of his knees with limping.
Patient 3, a first cousin of patients 1 and 2, was born at term. Immediately after birth he developed pneumothorax and was kept in hospital for 7 days. Growth and development have been slow, and he remains small at less than 10th percentile for height and weight. He has a chronic cough and has had reactive airway disease since infancy. He suffered multiple episodes of otitis media as well as pneumonia and 1 episode of urinary tract infection. He had visible tonsillar tissue as well as palpable lymph nodes.
Evaluation of the immune system
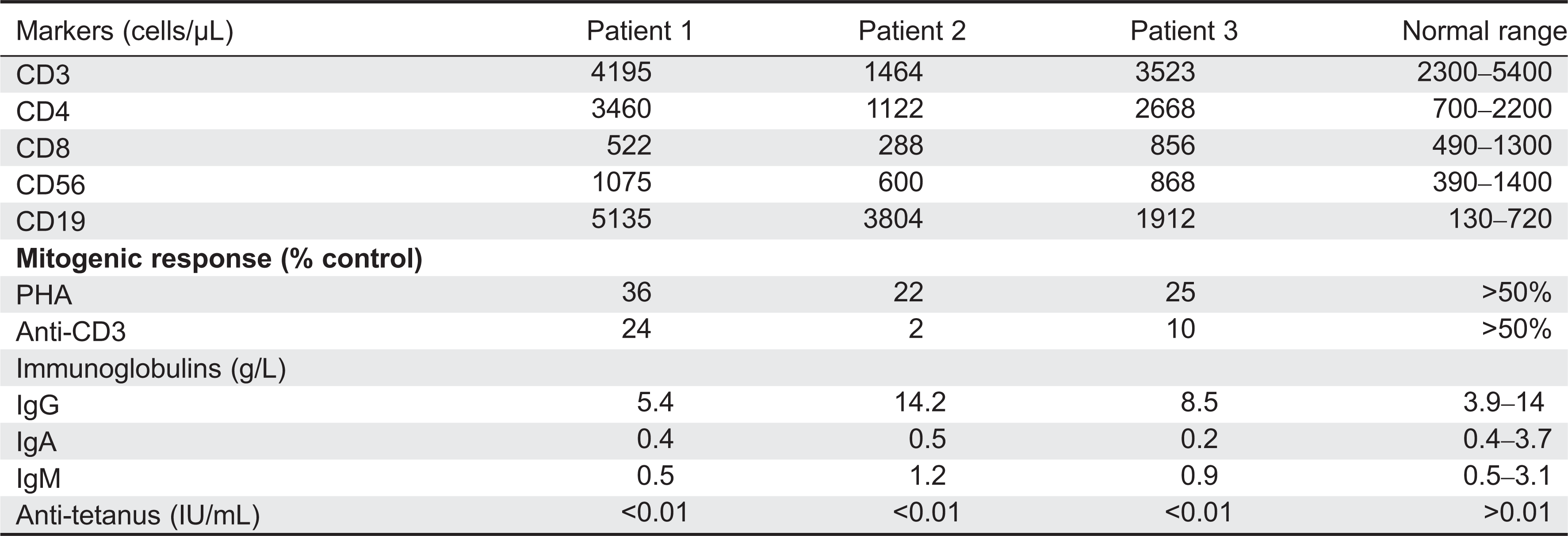
All patients displayed normal numbers of circulating white blood cells upon presentation. Immunophenotyping revealed that circulating lymphocyte numbers were normal to elevated in all 3 patients. (
Table 1). Although the number of circulating CD3
+ T cells was within normal range for age, CD4
+ T cells were somewhat increased, resulting in aberrant CD4:CD8 ratios of 4–10:1 (controls being 1.8–2.5:1). CD20 positive B-cell numbers were also significantly elevated, whereas CD56 positive natural killer cells were normal.
Despite the number of circulating T cells, in-vitro responses to PHA were less than 50% of normal controls and in-vitro responses to antigens including candida, zoster, simplex, and cytomegalovirus were completely flat (not shown). Total serum immunoglobulin levels were normal, but the ability to produce specific antibodies in vivo appeared to be significantly impaired (
Table 1).
Linkage analysis
Four family members, corresponding to the 2 male affected siblings and the parents, were analyzed using Illumina linkage panel chip (5913 single nucleotide polymorphisms (SNPs)). All of the 4 samples had a call rate greater than 98.9%. The self-reported gender and pedigree structure were confirmed by genotype data analysis using PLINK (
Purcell et al. 2007).
MERLIN (
Abecasis et al. 2002) was used for parametric linkage analysis. The minor allele frequency of the 4135 common SNPs present in both HapMap III and the Illumina linkage panel were estimated based on HapMap CEU
1 samples. Disease was assumed to be autosomal recessive with full penetrance as: 0.0, 0.1, and 1.0 (no mutated allele, 1 mutated allele, and 2 mutated alleles, respectively).
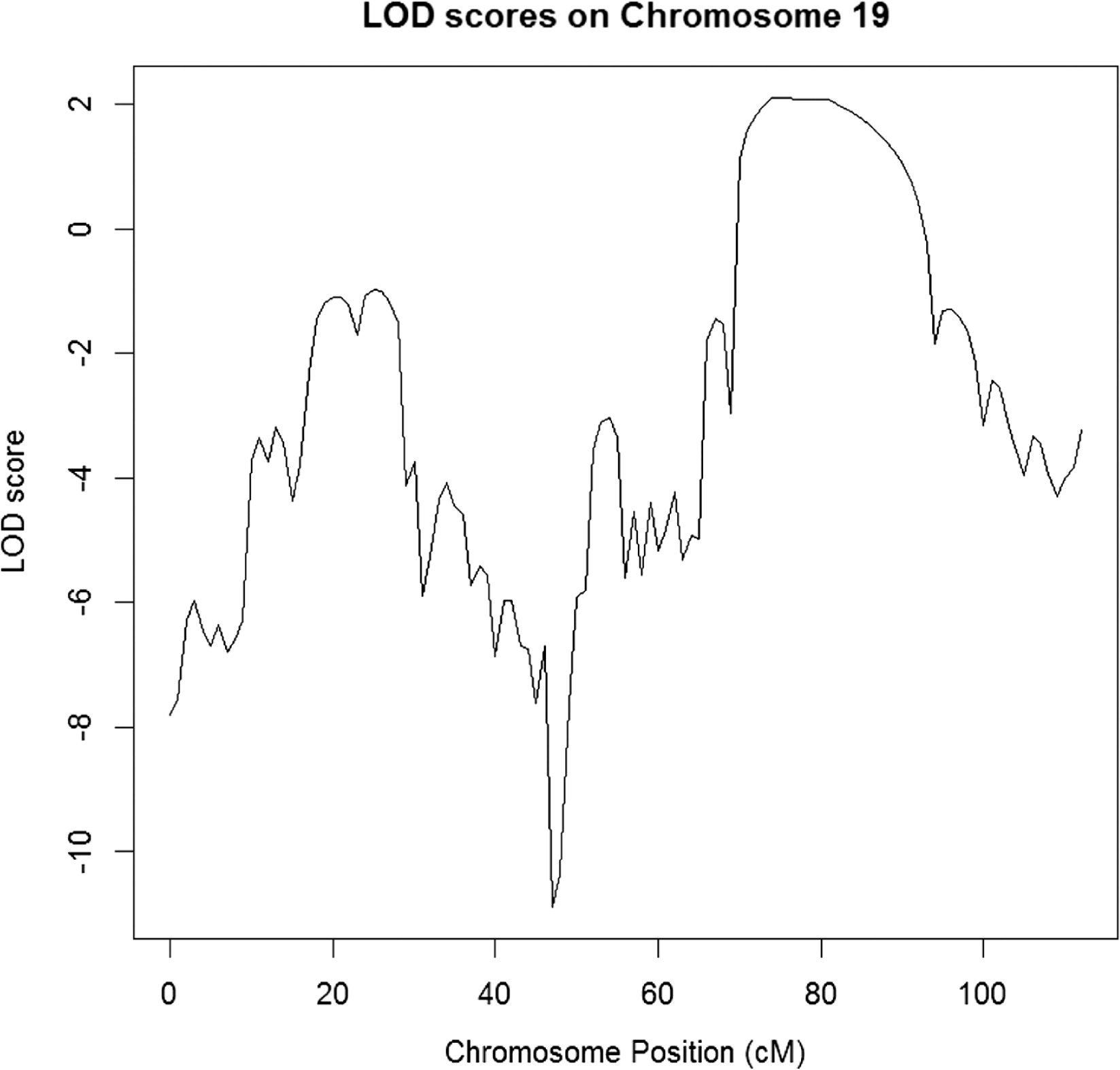
The parametric LOD score was calculated for a 1 cM grid along the chromosomes at disease allele frequency of 1%. Four chromosome regions were identified with a maximum LOD score larger than 2 (hg19 genomic coordinates) (
Figure 2):
Chromosome 3 (i): start = rs11720298, 70.9405 cM, 51 581 509 (hg19); end = rs1472653, 80.094 cM, 59 585 592 (hg19);
Chromosome 3 (ii): start = rs1317244, 120.54 cM, 112 053 565 (hg19); end = rs1052620, 144.177 cM, 136 574 521 (hg19)
Chromosome 18: start = rs736839, 70.0197 cM, 46 528 065 (hg19); end = rs1539964, 97.8493 cM, 67 625,285 (hg19)
Chromosome 19: start = rs1055099, 69.3765 cM, 43 979 589 (hg19); end = rs1017379, 91.09 cM, 52 457 306 (hg19)
Whole genome sequencing
The whole genome of patient 2 was sequenced using the Complete Genomics (Mountain View, Calif.) platform. The concentration of genomic DNA sample was measured with picogreen, and approximately 10 mg of DNA was submitted to Complete Genomics for whole genome sequencing. Complete Genomics employs high-density DNA nanoarrays that are concatemers of mate pair reads, each approximately 500 bp long. Base identification was performed using a non-sequential, unchained read technology known as combinatorial probe-anchor ligation (cPAL). Each mate pair includes 35 nt of genomic DNA sequence as well as adaptor sequences required for cPAL sequencing; the average mate gap length is 300 bp.
The resulting whole genome sequence had a fully called genome fraction (i.e., excluding sites with no-calls) of 96.7% and a fully called exome fraction of 98.3%. The genome fraction at weighted coverage ≥10 was 97.7% and the corresponding exome fraction was 98.8%; in particular, >60% of the genome was covered at >40× – summary statistics indicative of a high-quality sequencing run.
Disease variant identification
The search for the disease variant(s) was initiated by applying the following filters on single nucleotide variants, small indels, and small block substitutions: (
i) restrict to the 4 linkage peaks; (
ii) restrict to high-quality variants; we selected variants with the varQuality field of the Complete Genomics MasterVarBeta file equal to VQHIGH (corresponding to VAF, i.e., variable allele fraction model, quality score for both alleles equal or greater than 20 db for homozygous calls, and equal or greater than 40 for other calls); and (
iii) restrict to variants annotated as “coding” or “splicing” by either Complete Genomics’ annotations or in-house annotations based on Annovar (
Wang et al. 2010).
This procedure resulted in 50–500 variants per peak. We further restricted to homozygous variants, assuming a recessive model, and excluded synonymous variants. Variants were manually reviewed, taking into account: (
i) variant frequency in reference databases (1000 Genomes, NHLBI-ESP, Complete Genomics reference panel) (
Abecasis et al. 2010;
Tennessen et al. 2012) and presence in dbSNP137; (
ii) residue-level conservation (PhyloP score) and local sequence conservation (multi-species alignments as provided by the UCSC browser); (
iii) variant effect on coding sequence (e.g., missense, stop-gain, splicing); and (
iv) gene function, provided by Gene Ontology (
Ashburner et al. 2002), and associated phenotypes, provided by HPO/OMIM (OMIM descriptions as processed into a controlled vocabulary by the Human Phenotype Ontology) and MPO/MGI (Mammalian Phenotype Ontology annotations for mouse genes provided by Mouse Genome Informatics) (
Smith et al. 2005;
Robinson et al. 2008).
We identified a homozygous stop-gain in the gene RelB (chr19: 45,535,991). Both alleles had a high-quality score (variable allele fraction model: 111 and 981 db; equal allele fraction model: 50 and 52 db) and the variant was novel according to all reference databases (dbSNP, 1000 Genomes, NHLBI-ESP, Complete Genomics reference panel). Inspection of the locus in UCSC did not reveal any anomaly. In addition, NHLBI-ESP has not reported any stop-gain or splicing variant for RelB in over 5000 subjects, suggesting that the gene is heavily under negative selection against loss of function variants; it is specifically interesting that no loss of function altering the final exons were found either.
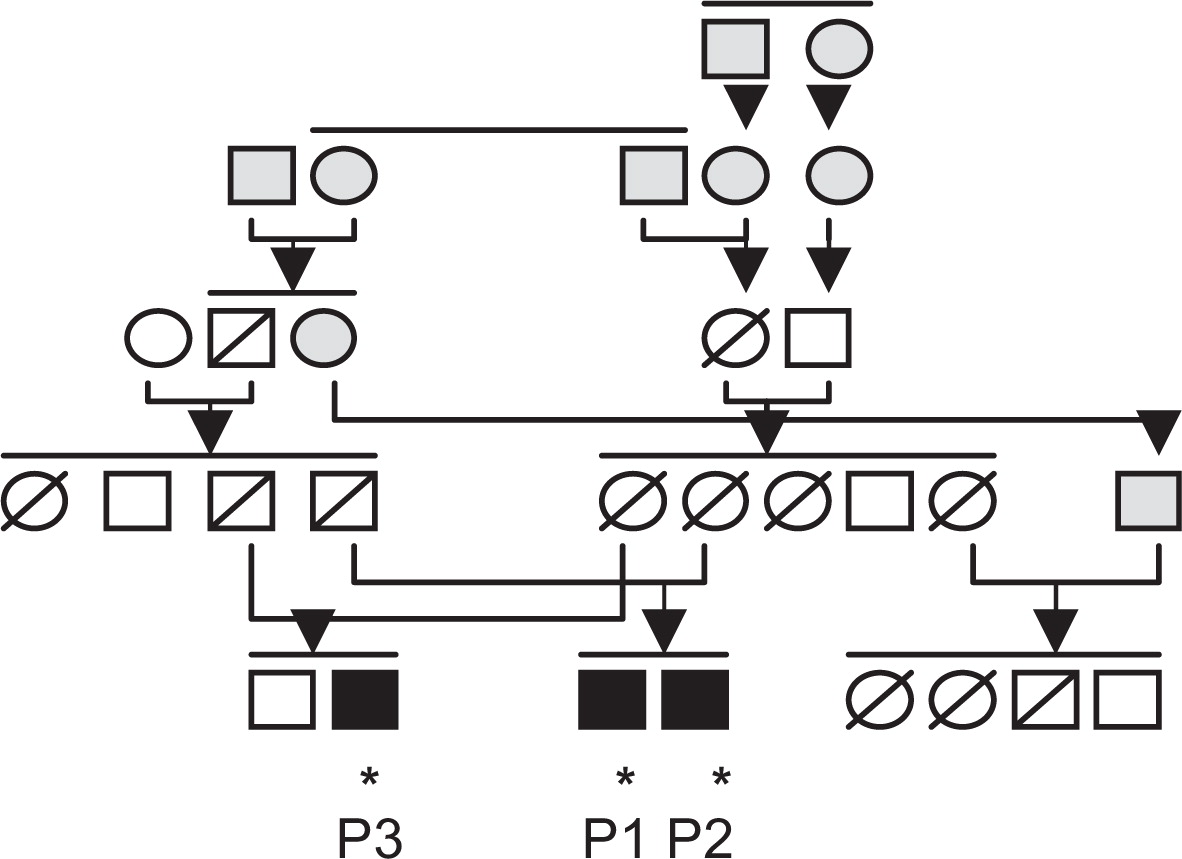
Subsequently, the stop-gain mutation was confirmed by Sanger sequencing and found to segregate with disease following the analysis of 21 family members (
Figure 3). All 3 affected children were homozygous for the
RelB mutation, all 4 parents were heterozygous, and unaffected siblings and close relatives were either heterozygous or carrying 2 wild-type alleles.
Quantification of RelB mRNA and protein levels
Relative levels of RelB mRNA were assessed by RT-PCR analysis in 2 patients to determine whether the stop-gain mutation affected RelB mRNA transcription. Total RNA was isolated from Epstein–Barr virus (EBV) transformed B-cell lines (normal control, patient 2, and patient 3) and reverse transcription carried out with SuperScript III RT and Oligo(dT)12–18 primer. Primer pairs were checked for specificity by standard PCR. Relative quantification was performed using the standard curve method. The normal control EBV cDNA was used as the calibrator and GAPDH as an endogenous control. Results were determined as the mean and standard deviation for 5 replicate samples within a single experiment. In both patients examined, RelB mRNA levels were found to be lower than normal control (
Table 2).
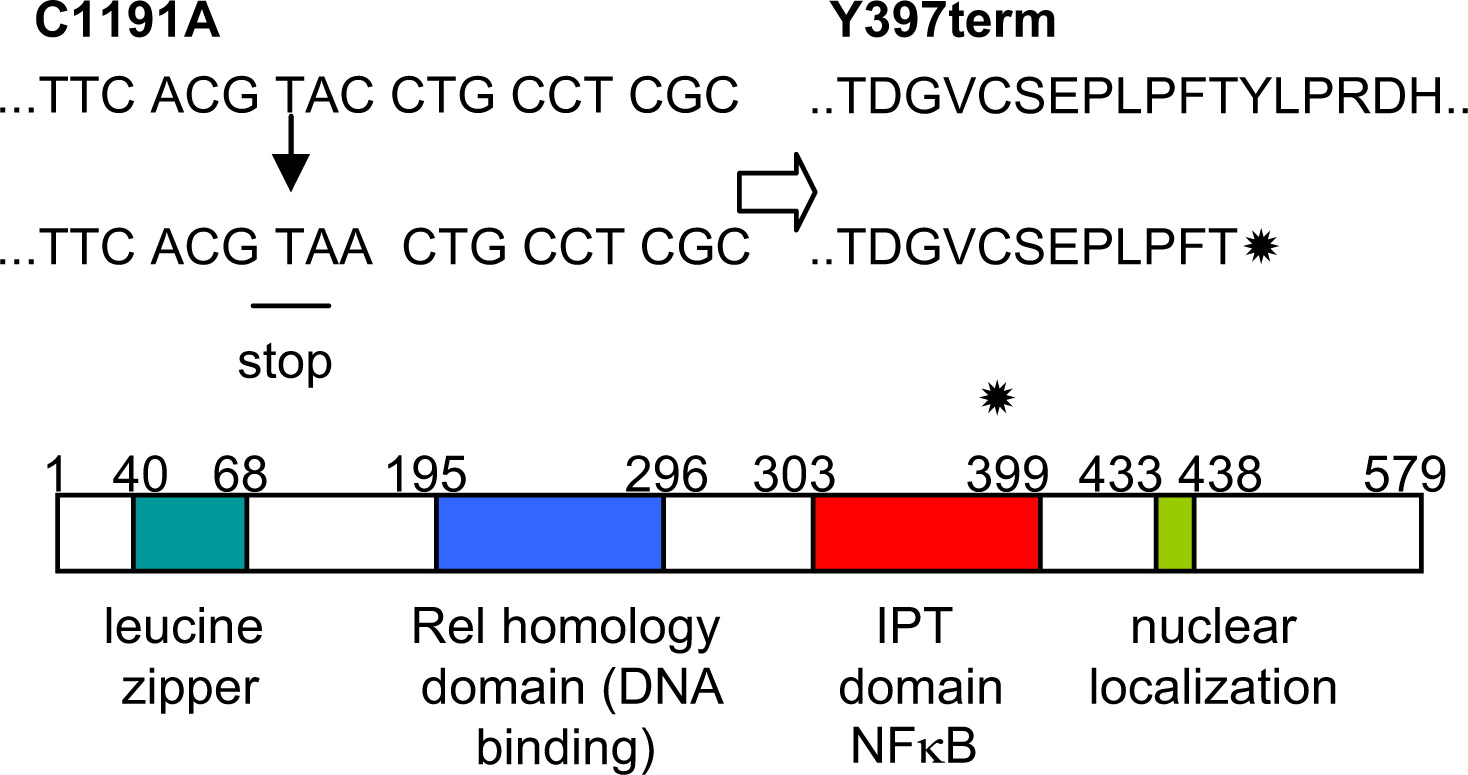
The mutation identified in
RelB created a premature stop codon that blocked translation of the last 3 coding exons, shortening the protein by approximately one-third (terminating at amino acid 396) and potentially creating a protein devoid of transcription activating properties, owing to the deletion of its C-terminal nuclear localization sequence (
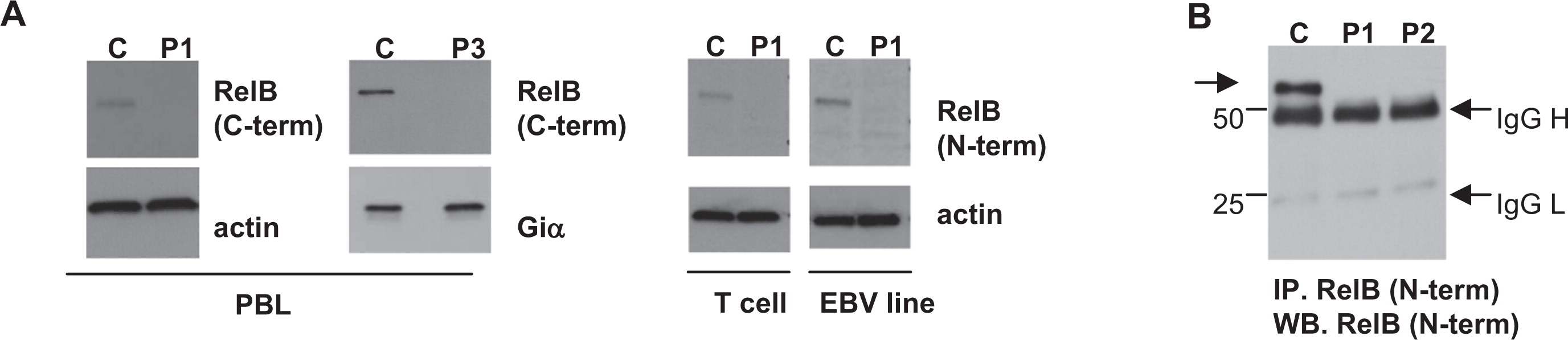
Figure 4). Western blotting with antibodies against the
RelB C-terminus, predicted to be deleted by the mutation, revealed no detectable RelB expression in the peripheral blood lymphocyte (PBL) of patient 1 and patient 3 (
Figure 5). Furthermore, antibodies against the retained
RelB N-terminal region also failed to detect any RelB protein in patient lysates. Enrichment by immunoprecipitation with anti-N terminus antibody and subsequent Western blotting similarly failed to detect any potential RelB fragment that would be coded by amino acids 1–396 if translated (predicted fragment, 43 kDa; wild type, 62 kDa) (
Figure 5B).
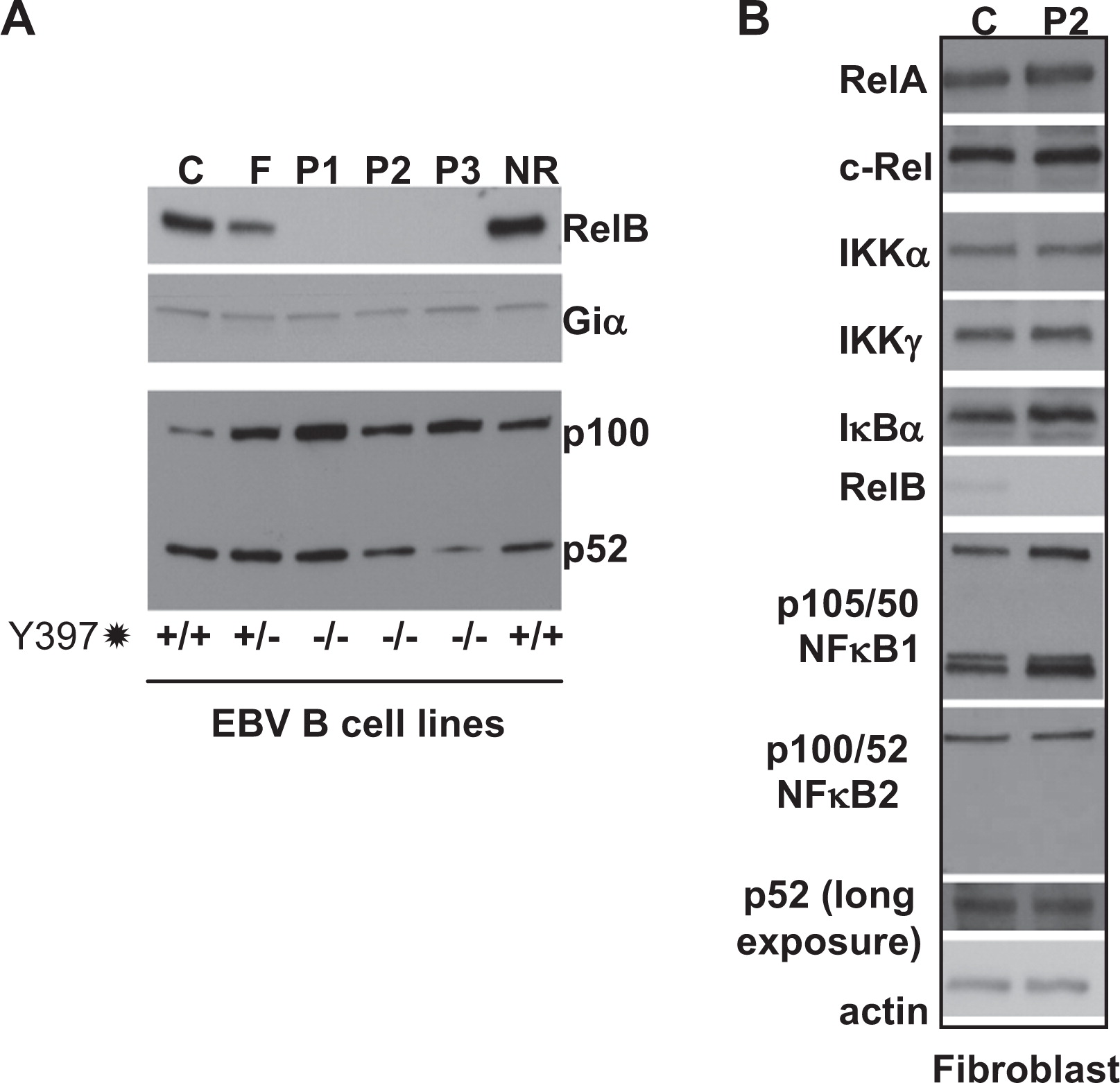
In contrast to RelB, all other components of the NFκB alternative and classical signaling pathways that appeared to be normally expressed were examined in patient EBV B cells and primary fibroblasts (
Figure 6). Thus, the premature stop codon appears to effectively ablate RelB expression in these patients, resulting in a “knockout” phenotype.
Discussion
We describe here a novel type of CID associated with a mutation in the
RelB gene. Patients presented with repeated infections and failure to thrive, hallmarks of severe combined immunodeficiency (SCID). As these patients have normal to increased (yet dysfunctional) numbers of circulating T cells, they would be classified as leaky SCIDS or a CID. Occasionally, these types of patients may have autoimmune manifestations such as Omenn syndrome or colitis, yet management and outcome are similar to SCID in most cases (
Roifman et al. 2012).
Patient consanguinity facilitated linkage analysis, several clear peaks with significant LOD scores were identified that ultimately helped pinpoint a homozygous mutations in RelB on chromosome 19. This premature stop codon mutation leads to decreased levels of RelB mRNA, presumably due to message instability. Furthermore, the RelB protein was undetectable in patient lymphocytes, suggesting that any potential fragment that may be translated is also unstable and subsequently degraded.
RelB −/− knockout mice die young of overwhelming inflammation in multiple organs owing to a T-cell dependent infiltration of tissues by myeloid lineage cells (although the exact mechanism is unclear) (
Weih et al. 1995,
1996). In contrast, humans with RelB deficiency do not develop this T-cell driven disease, but rather suffer repeated infections and persist with chronic lung and gastro-intestinal symptoms. Patients with defects of
NEMO or
IκBα in the related NFκB classical pathway may present with anhidrotic ectodermal dysplasia; however, this was not seen in our patients. RelB deficient patients did not display atypical mycobacterial susceptibility either, as observed in classical pathway deficiencies.
Invariably, all 3 patients had failure to thrive, a typical manifestation of combined immunodeficiency. Evaluation of the immune system revealed dysfunctional T cells that were unresponsive to mitogens or antigens in-vitro, and humoral immunity was also severely affected, with a failure to respond to childhood vaccines. Ultimately, symptoms were severe enough to warrant treatment with hematopoietic stem cell therapy.
The fact that RelB, a member of the alternative pathway of NFκB activation, appears so critical for the function of the immune system is surprising. RelB activity has been reported in signaling pathways downstream from only a few receptors: BAFF-R, CD40, TWEAK, and Lymphotoxin-β-R, suggesting that its deletion might have caused minimal phenotype. It is likely that the immune defects apparent in RelB-deficient humans are due to a wider effect of RelB on the classical NFκB pathway (involving RelA and c-Rel) through cross-regulation of activation and expression in addition to the function of RelB within the alternate pathway.