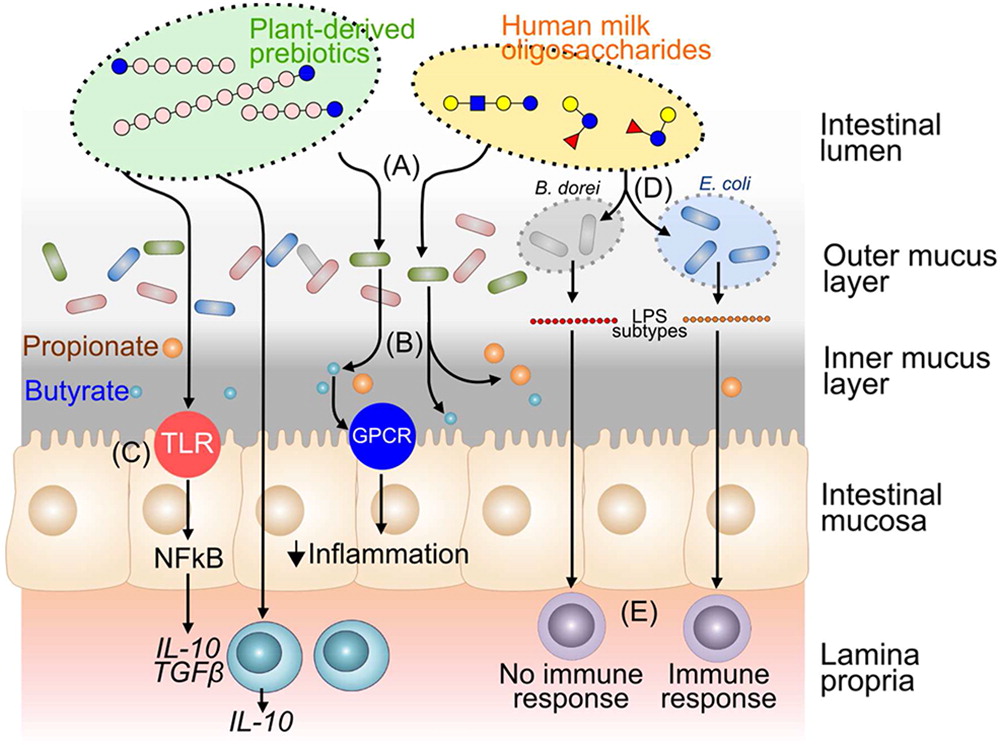
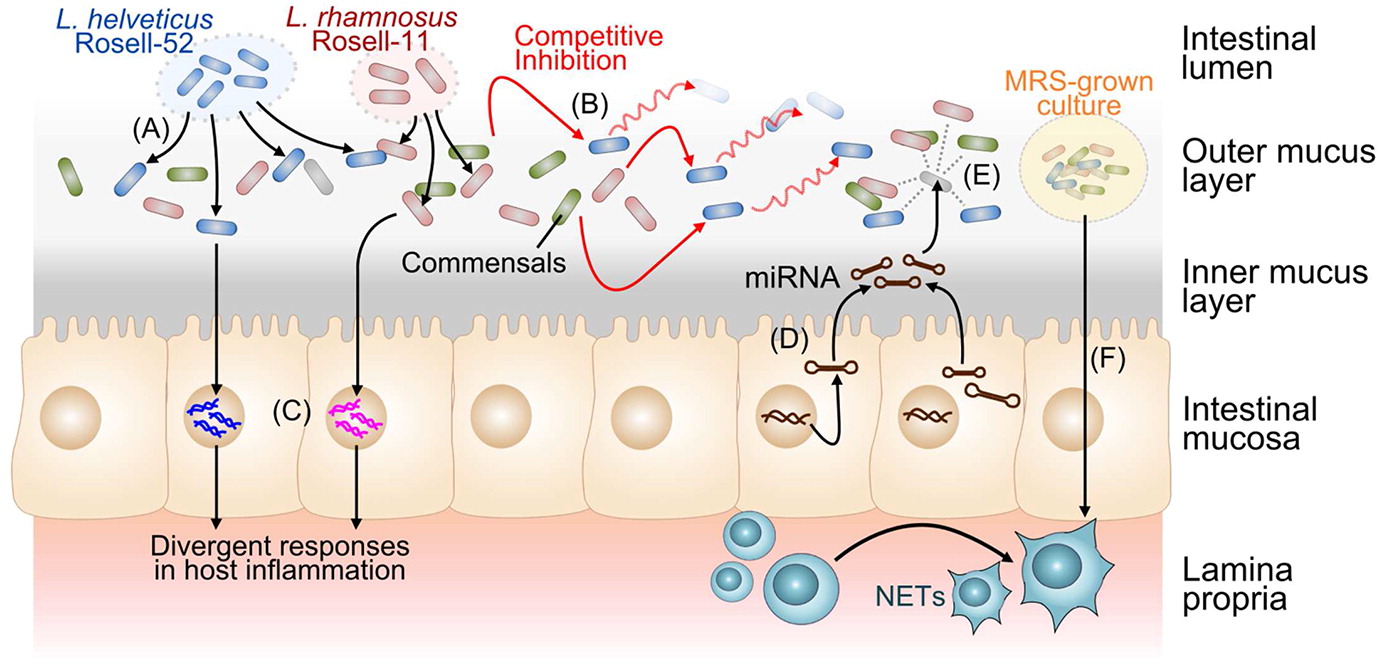
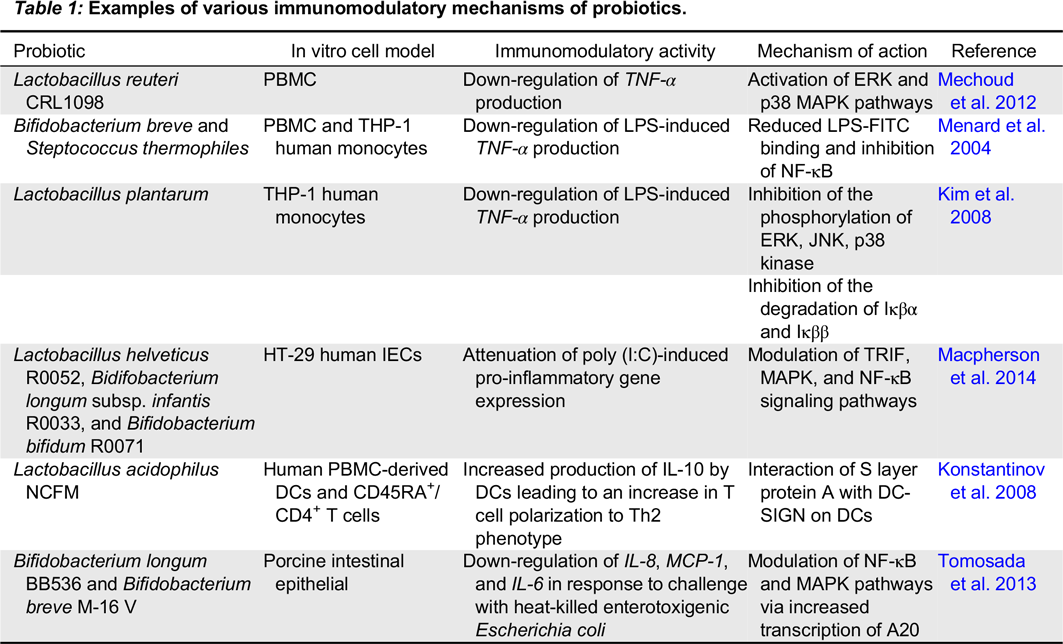
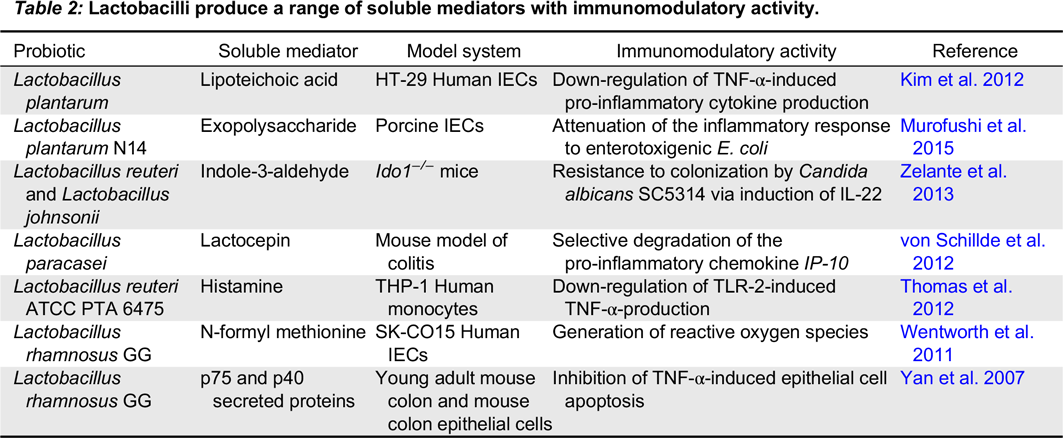
Numerous studies have explored interactions between probiotic bacteria and other cell types involved in both innate and adaptive immunity. While many recent studies focus on the role of probiotics in immune homeostasis, some probiotic strains interact with monocytes and macrophages to elicit an immunostimulatory effect. For example, the probiotic mixture VSL#3 activates NF-κB in macrophages resulting in increased production of IL-6 and IL-12, an effect mediated by TLR-9 recognition of DNA derived from these probiotic strains (
Rachmilewitz et al. 2004).
L. rhamnosus strain GG induces NF-κB activation and expression of
IL-6 in human peripheral blood mononuclear cells (
Miettinen et al. 2000) and decreases LPS Additionally, LGG induces TNF-α production by human THP-1 macrophages by inducing STAT3 activation. This leads to inhibition of c-Jun-N-terminal kinase (JNK) activation and induction of granulocyte-Colony Stimulating Factor (G-CSF), which, in turn, inhibits TNF-α production (
Kim et al. 2006). This effect is also mediated by culture supernatants from
L. rhamnosus strain GG and
L. rhamnosus GR-1 (
Kim et al. 2006). Supernatants derived from
L. rhamnosus strain GG also increase both TNF-α and IL-10 production by RAW265.7 murine macrophages, with effects mediated through TLR-2 and NOD2 (
Harb et al. 2013). Conditioned media from
L. rhamnosus strain GG upregulates
TLR-1,
-4,
-5,
-6,
-7,
-8, and
-9 expression by human blood-derived macrophages and dendritic cells, as well as MHC class II expression by macrophages, suggesting an impact on both innate recognition and antigen presentation efficiency by these APC (
Fong et al. 2016).
Probiotics also modulate macrophage phenotype and activity. Tissue macrophages exist as a heterogeneous population, broadly categorized into M1 and M2 macrophages. M1, or classically activated macrophages, are closely associated with the development of many inflammatory diseases, and secrete pro-inflammatory cytokines such as IL-1β, IL-6, and TNF-α. In contrast, M2 macrophages, or alternatively activated macrophages, play a regulatory and anti-inflammatory role through the secretion of the regulatory cytokines IL-10 and IL-1Ra. Macrophages exhibit phenotype plasticity, as they are able to switch between M1 and M2 depending upon external stimuli (
Martinez and Gordon 2014). Treatment of non-differentiated macrophages with the probiotic mixture VSL#3 results in an increase in the regulatory cytokines
IL-1Ra,
IL-13, epidermal growth factor (
EGF), fibroblast growth factor-2 (
FGF-2), transforming growth factor
(TGF)-α, vascular endothelial growth factor (
VEGF), as well as pro-inflammatory cytokines, including
IL-12 and
TNF-α suggesting differentiation into the immunoregulatory M2b macrophage phenotype (
Isidro et al. 2014). Gram-negative bacteria, such as species of
Escherichia and
Salmonella, are more potent inducers of M1 activation than Gram-positive bacteria, with strain-specific effects observed (
Christoffersen et al. 2014). A fermented milk product containing
L. paracasei FT700 induces differentiation of monocytes into immunoregulatory macrophages (
Tulini et al. 2015), suggesting a possible mechanism of action for a probiotic functional food. A comparison of the impact of 5 strains of heat-killed probiotics and their secreted proteins on LPS-induced TNF-α and IL-6 production by THP-1-derived M1 and M2 macrophages reveals differential effects, depending on formulation and on macrophage subtypes (
Habil et al. 2011). For example, secreted proteins and heat-killed strains of
Bifidobacterium and
Lactobacillus suppress IL-6 production by M2 macrophages, but differ in their impact on M1 macrophages. Production of the pro-inflammatory cytokine TNF-α is augmented by both formulations in CD14
hi M1 macrophages, reflecting the complexity of probiotic-APC interactions
in vivo, and demonstrating the importance of considering probiotic formulations in potential therapeutic applications.