The effects of STAT1 dysfunction on the gut
Abstract
Introduction
Methods
Patient enrollment
Clinical features of patients
Genetic analysis
Histology analysis
Immunohistochemistry for T-lymphocyte markers CD3, CD4, and CD8
Multilabel immunofluorescence and confocal microscopy
Lymphoblast culture, cell growth, and viability analysis
Protein expression analysis
Results
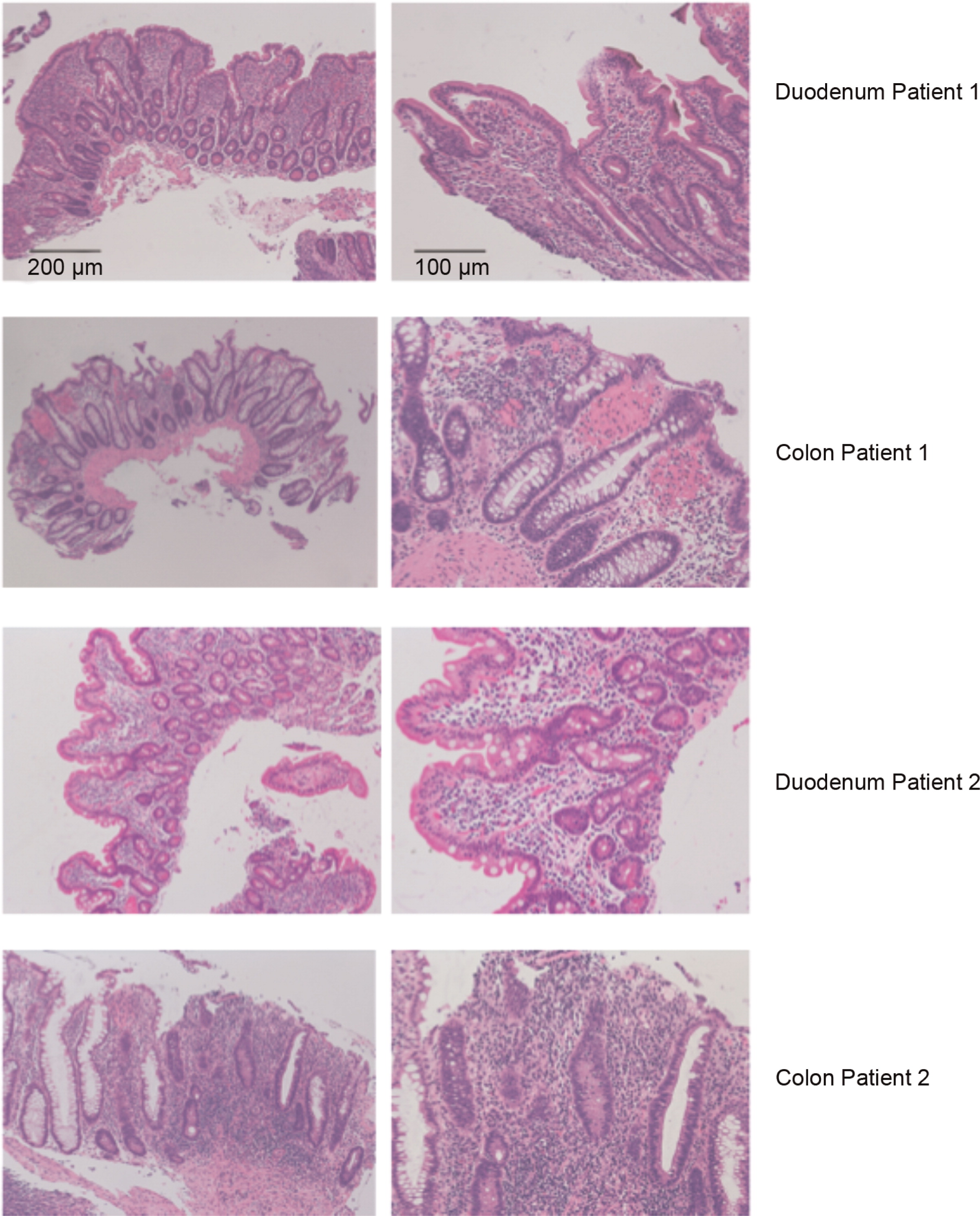
Histopathology analysis of patients with mutations in STAT1
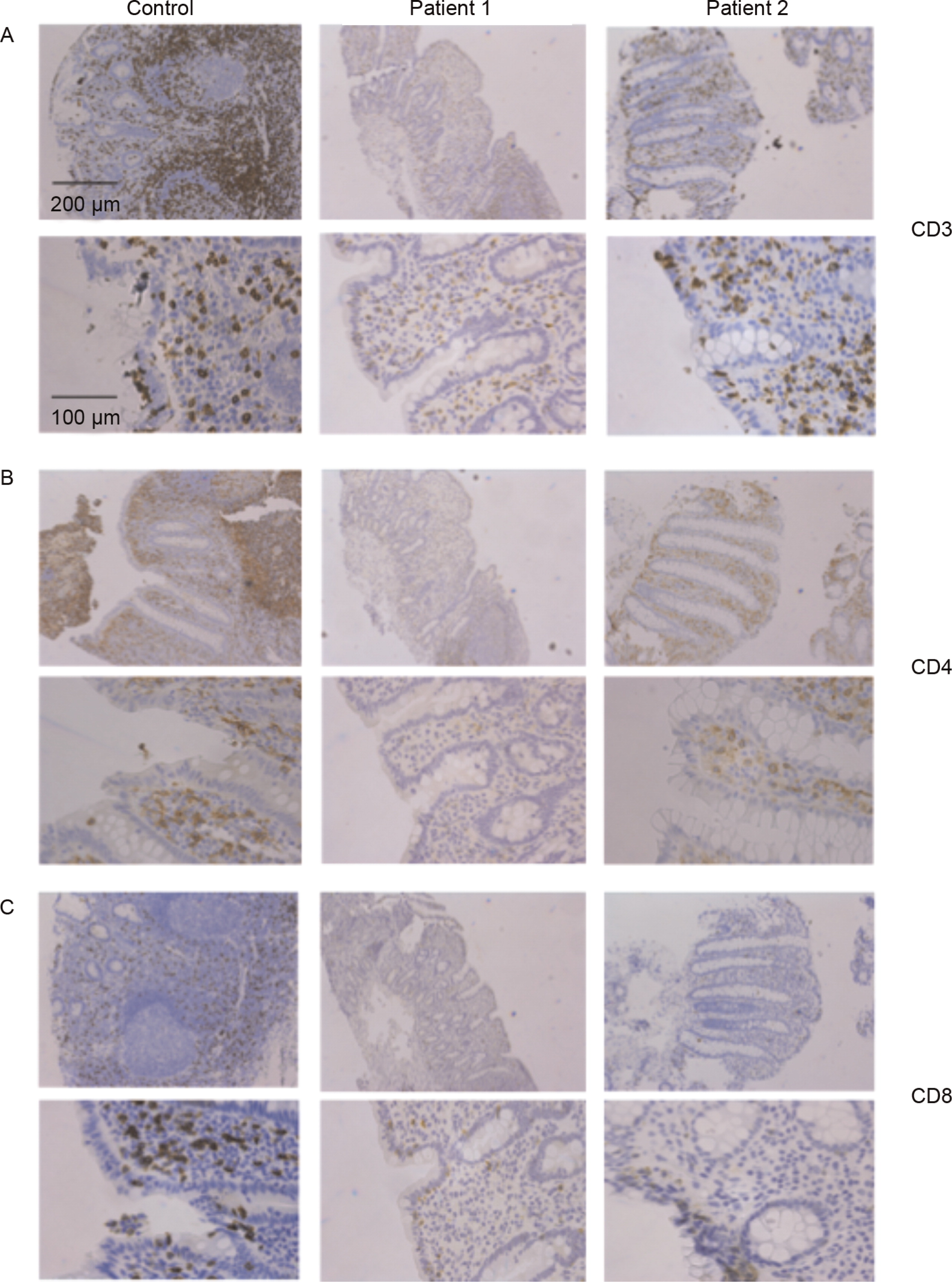
Lymphocyte populations in patients’ intestinal biopsies
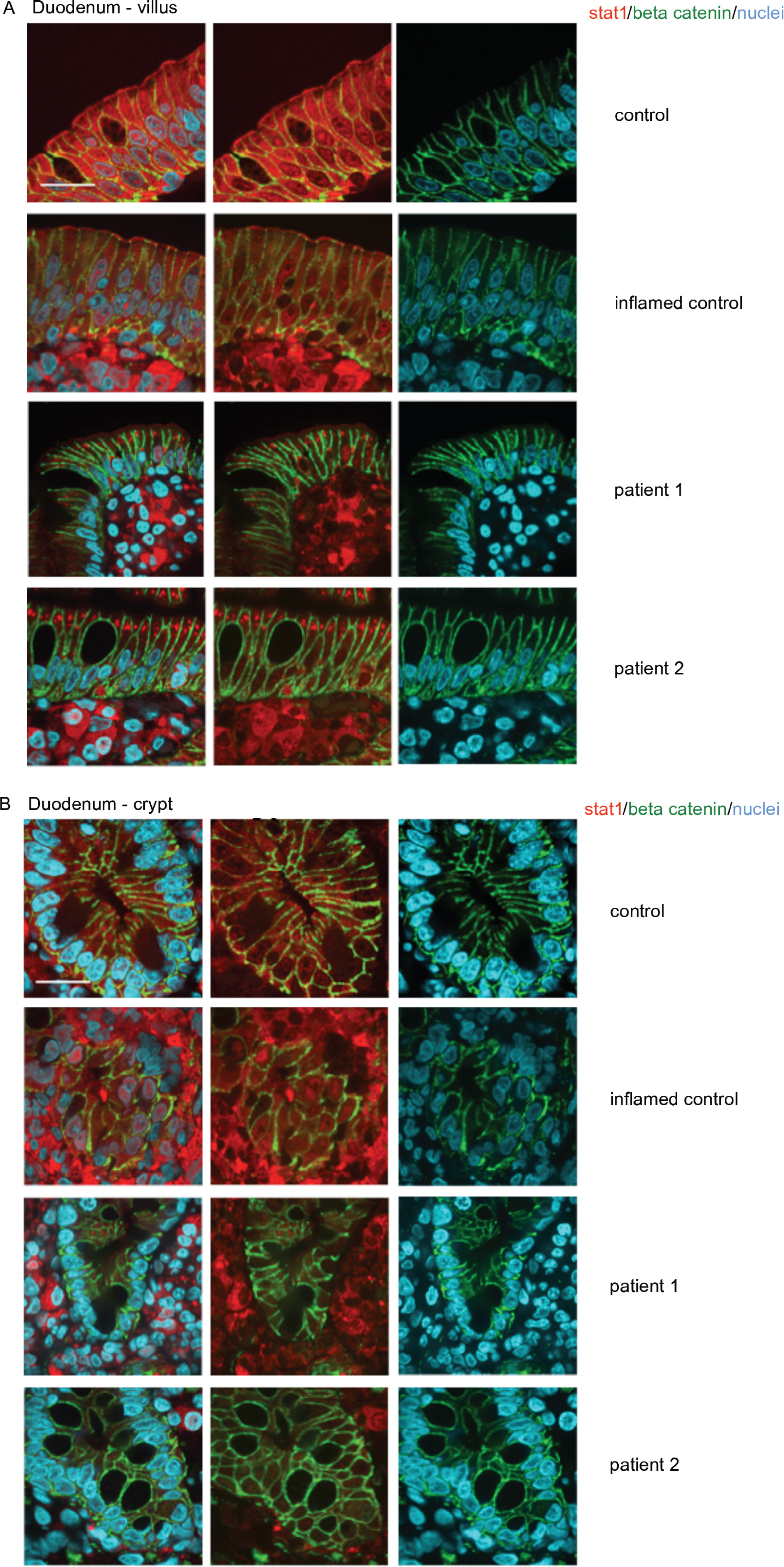
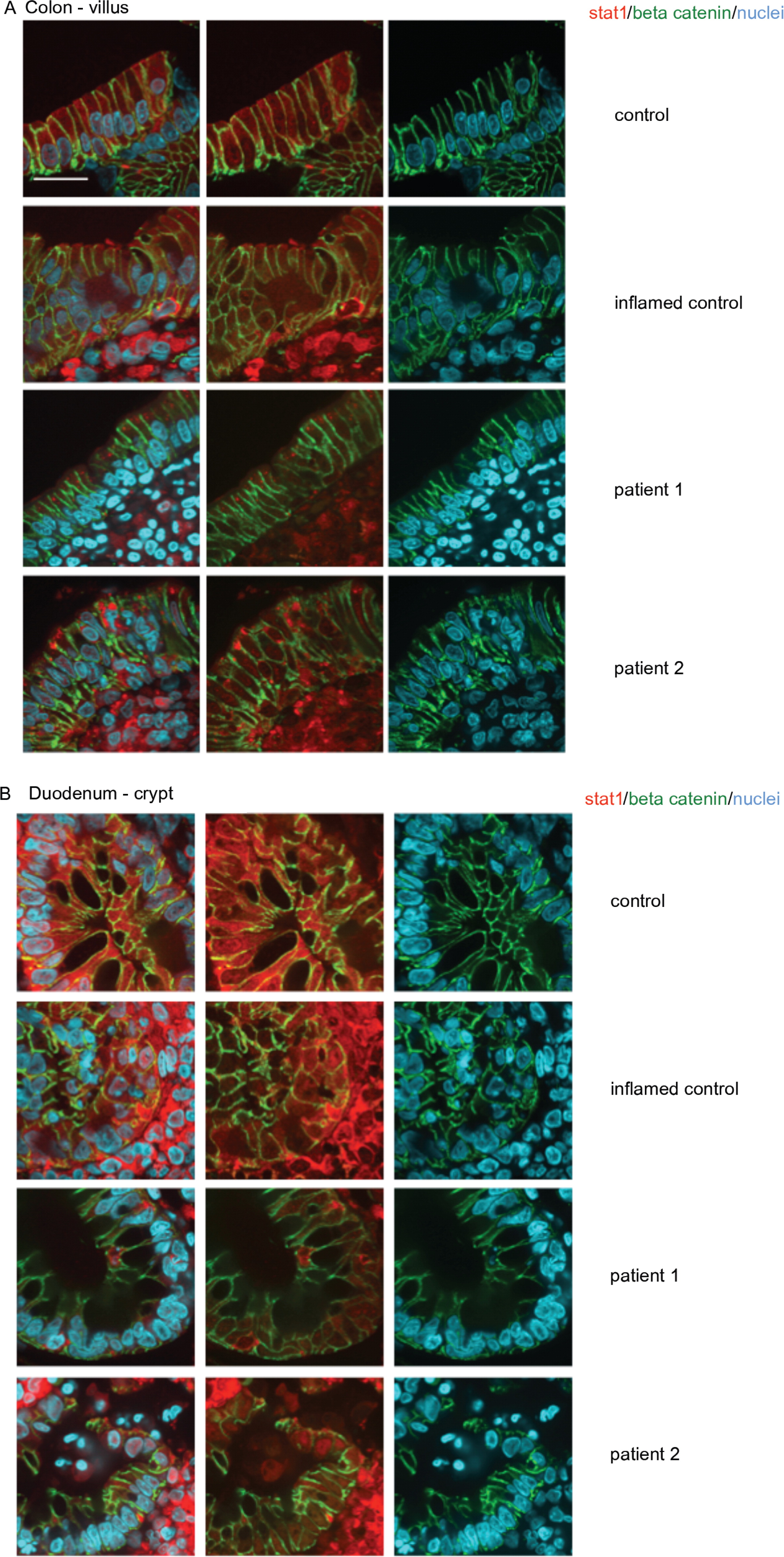
Expression of total and activated STAT1 in enterocytes
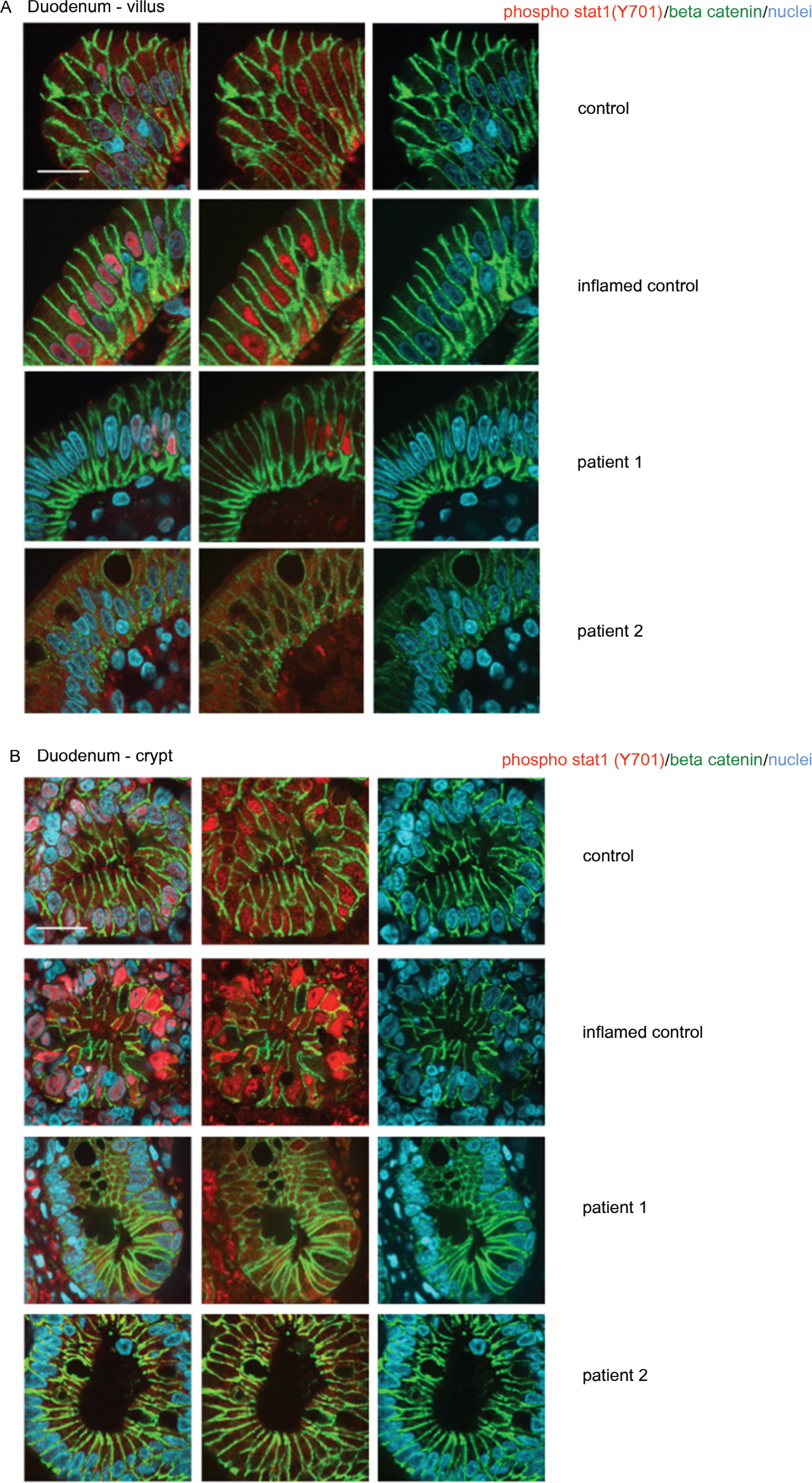
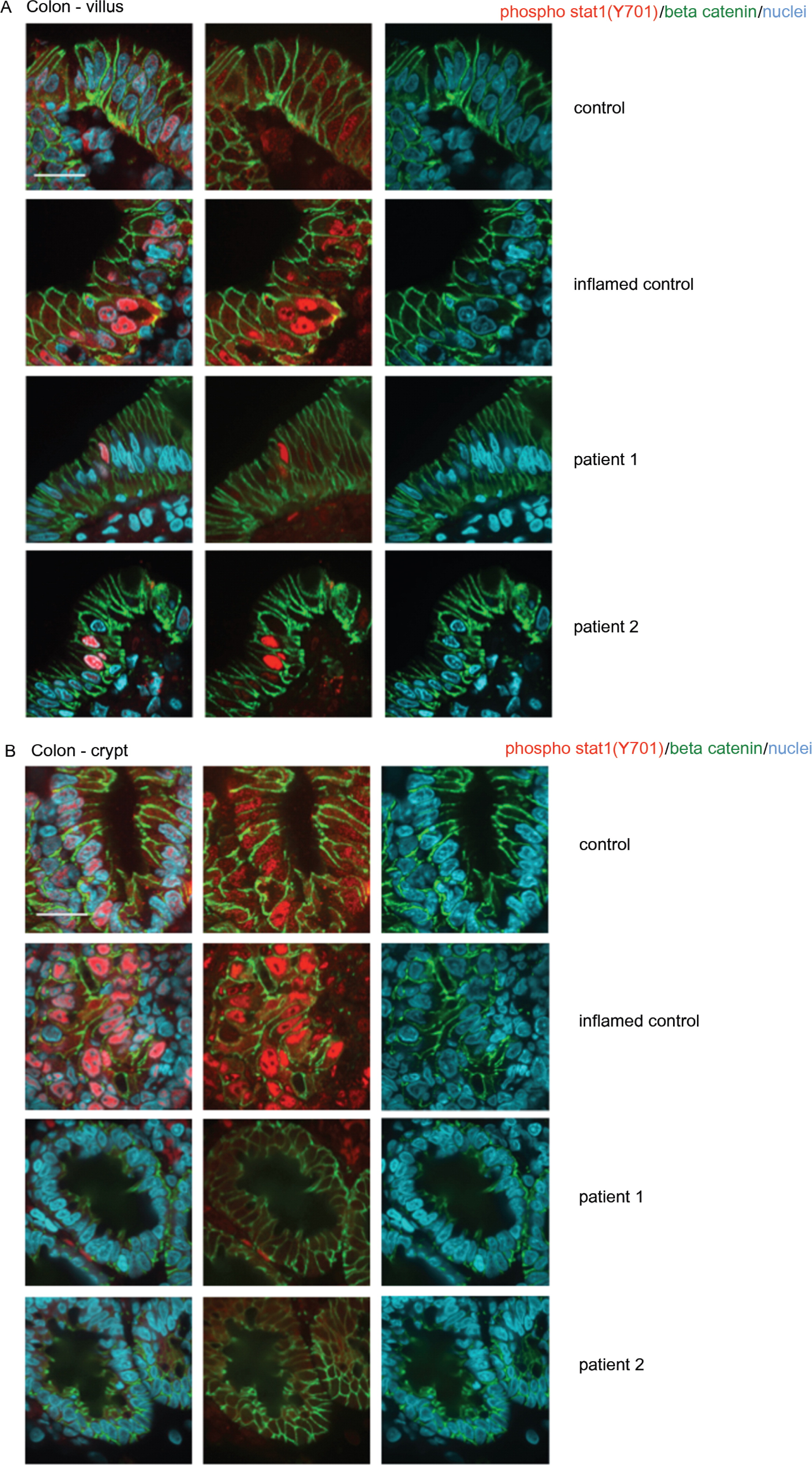
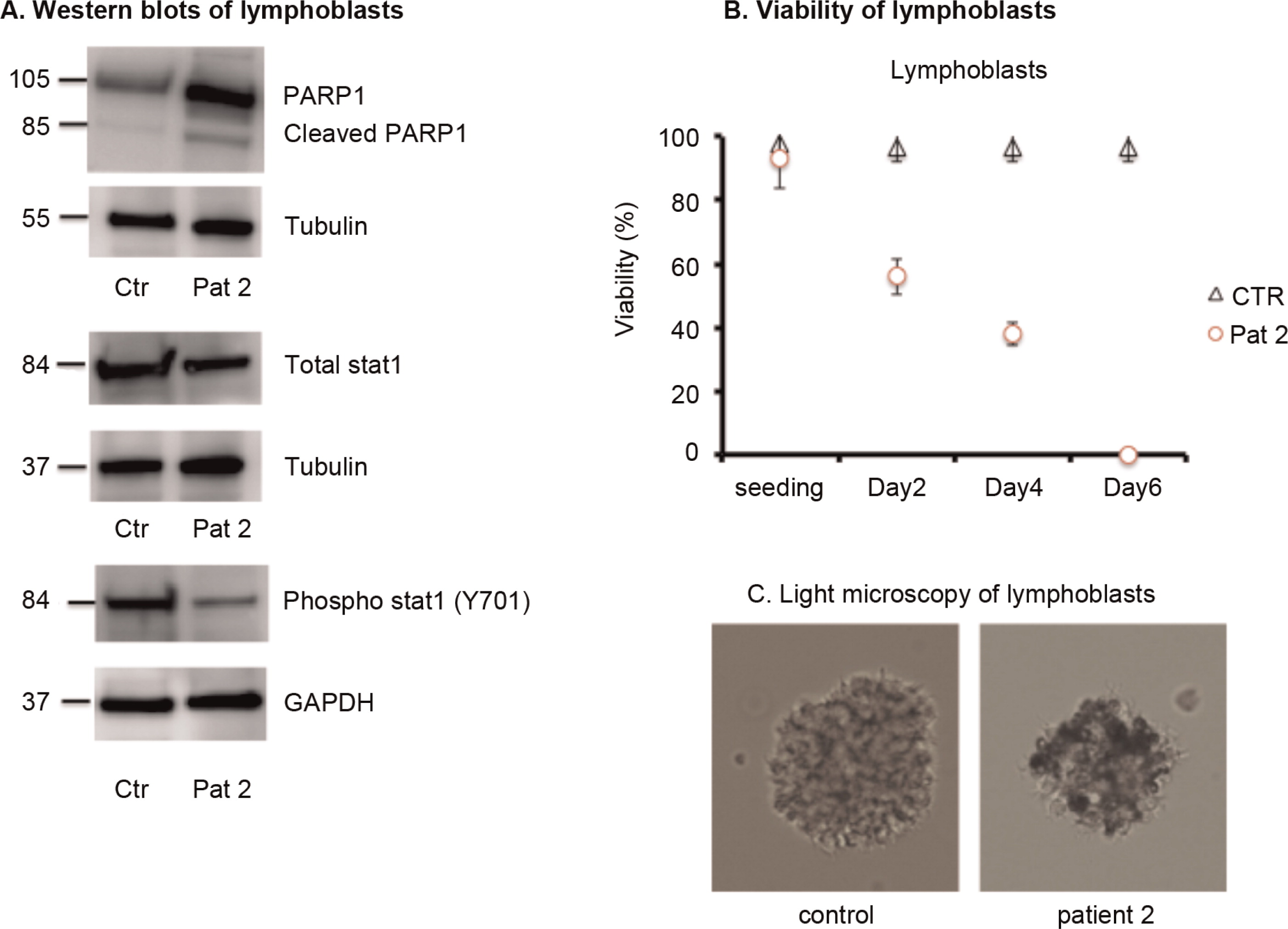
Analysis of patient lymphoblasts
Discussion
Acknowledgments
REFERENCES
Information & Authors
Information
Published In

History
Authors
Metrics & Citations
Metrics
Other Metrics
Citations
Cite As
Export Citations
If you have the appropriate software installed, you can download article citation data to the citation manager of your choice. Simply select your manager software from the list below and click Download.
There are no citations for this item
View Options
View options
Login options
Check if you access through your login credentials or your institution to get full access on this article.


