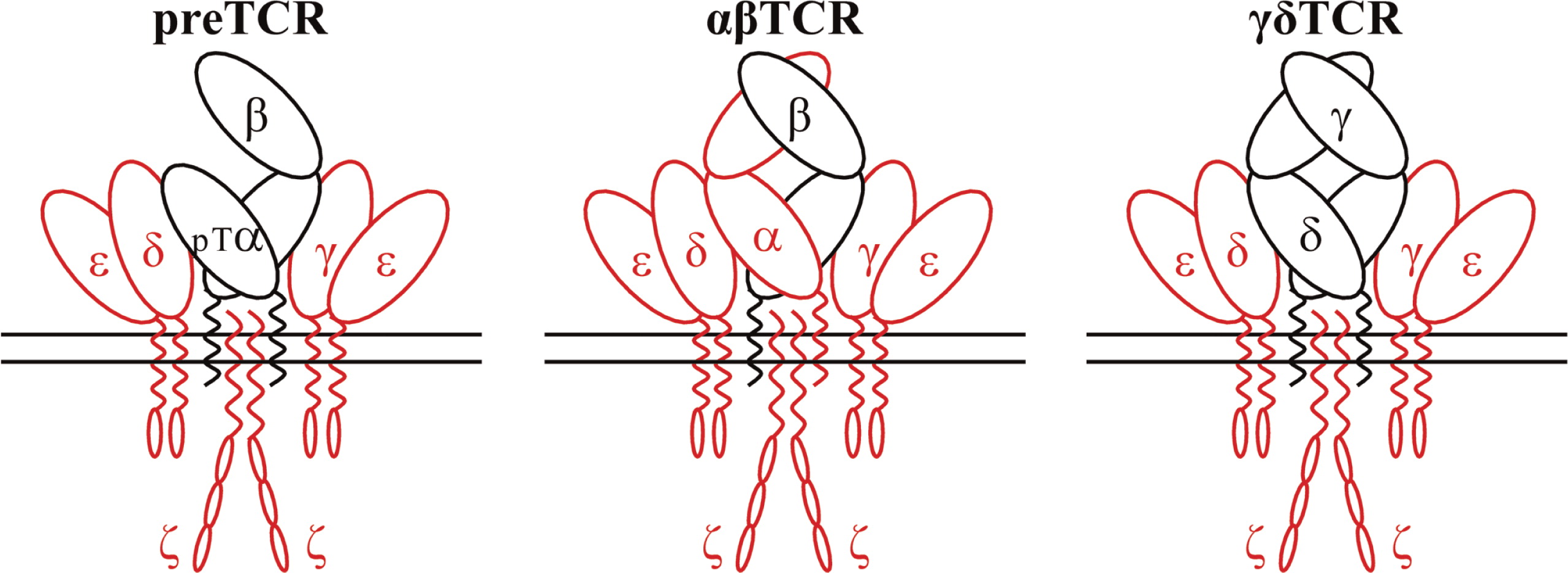
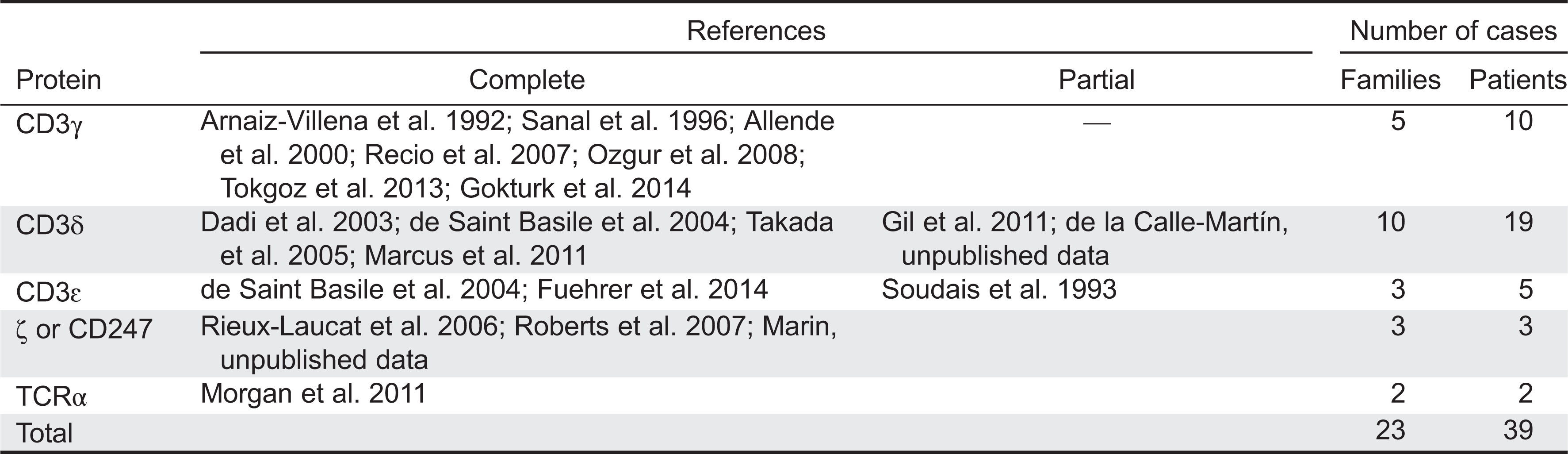
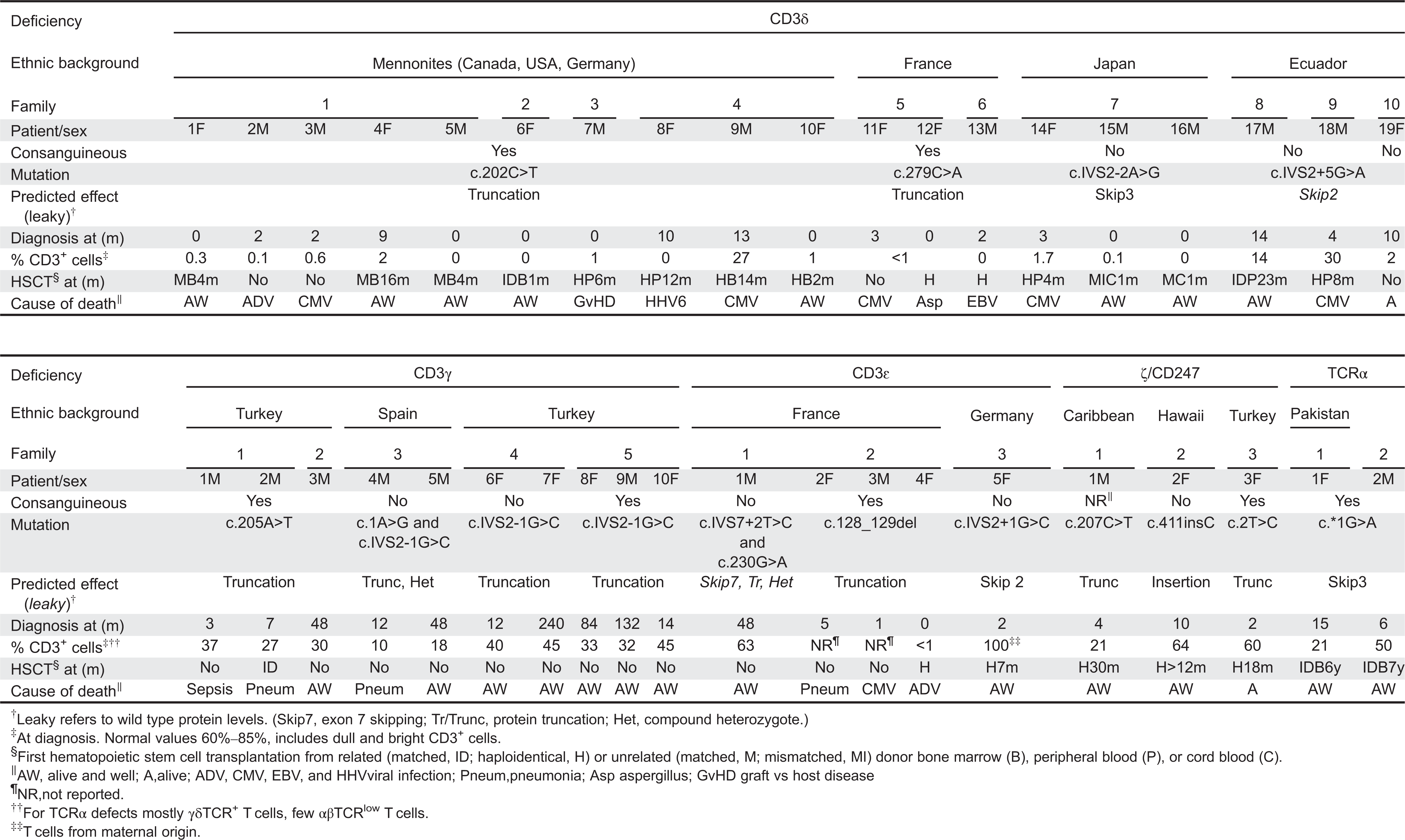
T-cell subset definition and lymphopenia
TCRID may have no T cells and are thus properly called T
−B
+NK
+. But often some T cells are present (T
±B
+NK
+), although their identification using CD3 as an extracellular marker can be difficult owing to their reduced TCR expression. Indeed, some authors have reported T
−B
+NK
+ phenotypes despite the presence of abundant CD4
+ and CD8
+ lymphocytes expressing very low levels of surface CD3 (
Roberts et al. 2007). As intracellular CD3 is not normally used for screening TCRID, we used extracellular CD4 and CD8 to identify T cells in TCRID patients. We have shown that essentially all CD4
+ and CD8
bright cells within the lymphocyte gate are αβ T cells and, conversely, most double negative cells (DN, CD4
−CD8
−) in the same gate are γδ T cells (
Muñoz-Ruiz et al. 2013). CD8
dull lymphocytes are mostly NK cells. For comparative purposes, we have considered CD3
+ lymphocytes in
Table 2 because they are widely reported, although it is likely an underestimation of the true number of T cells. Chimerism should be excluded using standard fingerprinting procedures, to avoid for instance counting the mother's T cells, as reported recently in a new case of CD3ɛ deficiency (
Fuehrer et al. 2014).
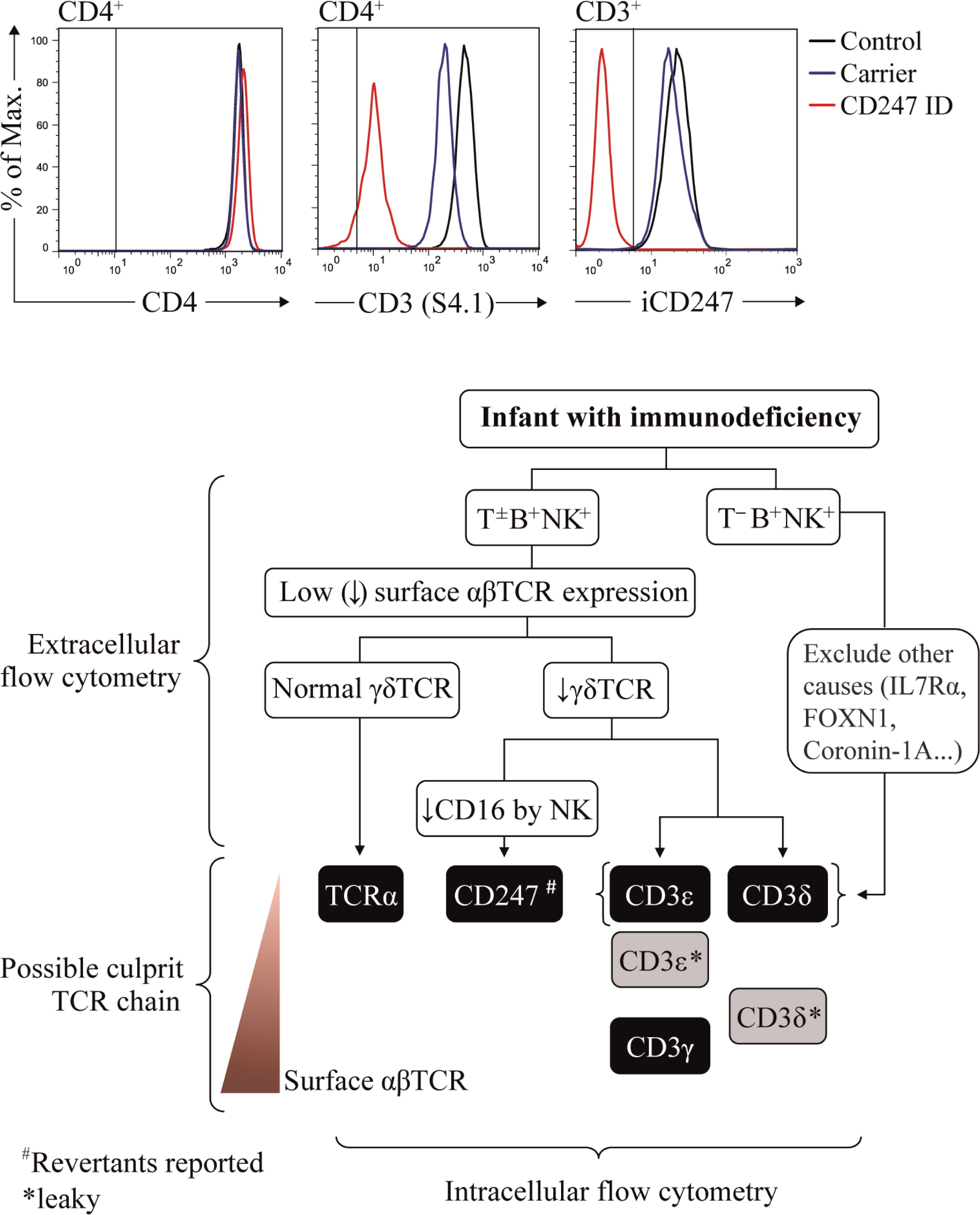
Using these criteria, most patients with complete defects show selective peripheral blood T lymphopenia, either T−B+NK+ or T±B+NK+ depending on the affected TCR chain, which is a first clue to the molecular basis of the pathology. T−B+NK+ has been associated with complete CD3ɛ or CD3δ defects, with less than 2% peripheral blood T cells. T±B+NK+ has been reported in complete CD3γ, TCRα, or CD247 defects. All partial TCRID show a T±B+NK+ or even a T+B+NK+ immunophenotype.
TCRID are best diagnosed if both αβ and γδ T cells are studied, as shown for complete TCRα deficiency (
Morgan et al. 2011) and for partial CD3δ deficiency (
Gil et al. 2011), both of which shared a Tαβ
−γδ
+B
+NK
+ immunophenotype despite their disparate molecular basis.
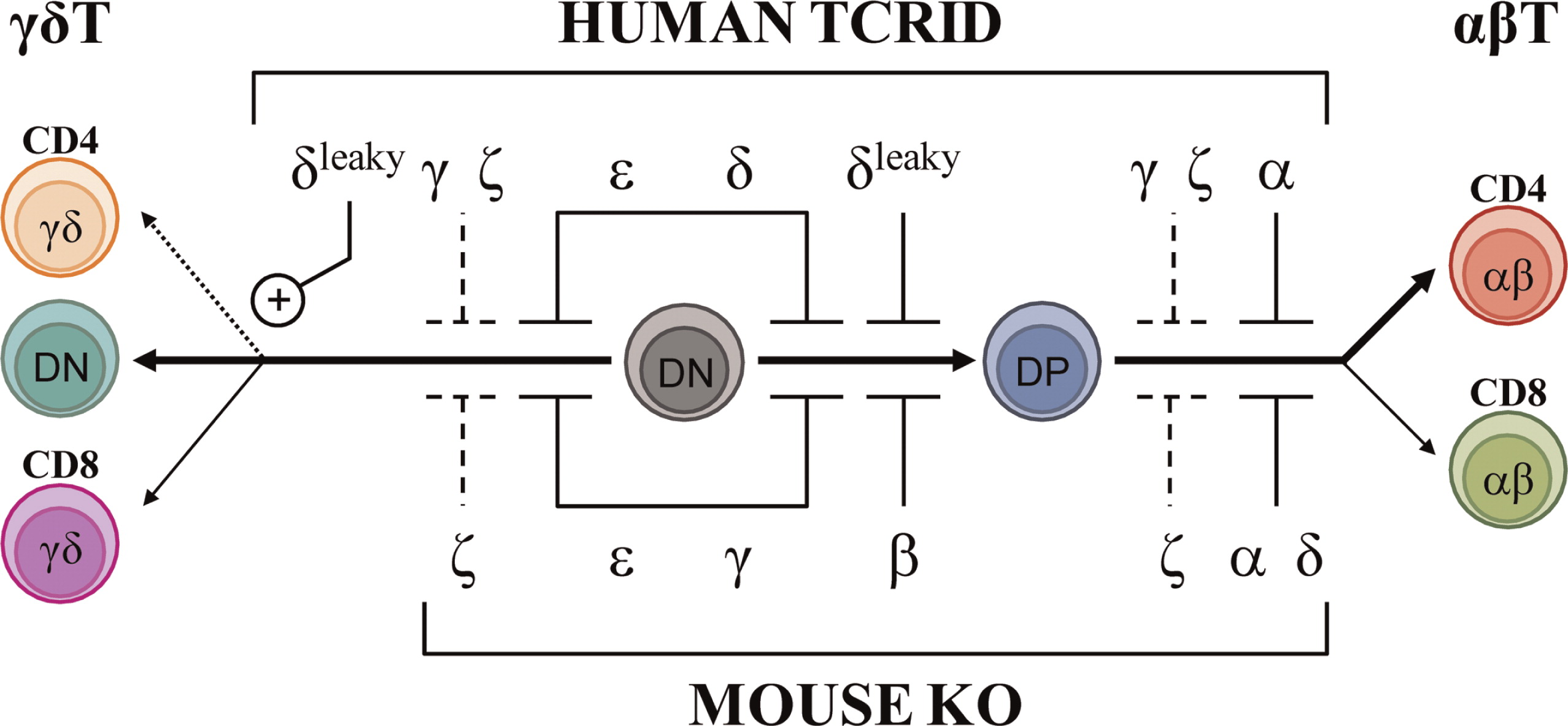
Figure 2 (top) summarizes the effects of TCRID mutations in T-cell development.
Surface TCR expression in patients’ lymphocytes
We have listed the monoclonal antibodies used in our lab for surface staining of peripheral blood lymphocytes from possible TCRID patients (
Table 3, extracellular).
We start with just UCHT-1 (anti-CD3ɛ), 11F2 (anti-γδTCR), and any anti-CD4 and anti-CD8α antibodies, and in a different staining IP26 (anti-αβTCR) and 11F2 (anti-γδTCR). Within a subset (for instance, CD4
+ lymphocytes), TCR Mean Fluorescence Intensity is evaluated with any of those CD3- or TCR-specific antibodies in comparison with that of an age-matched healthy control (
Figure 3, top center). The expression of CD4 itself can serve as an internal control (
Figure 3, top left). We normally use a range of CD3-specific monoclonal antibodies, such as those listed in
Table 3 (extracellular), to confirm any TCR expression defect.
Family members are not appropriate controls, because in some cases they also show (partial) TCR expression defects that may underestimate the patient's defect (see
Figure 3 and
Muñoz-Ruiz et al. 2013, top center and “Surface TCR expression in carriers’ lymphocytes”). If no TCR expression defect is observed, TCRID can be excluded. If a TCR expression defect is ascertained, both αβ and γδ T cells should be analyzed. If αβ T cells (defined as CD4
+ and (or) CD8
bright) show impaired TCR expression and γδTCR
+ show normal surface TCR expression, TCRα deficiency is likely. If αβ and γδ T cells share a surface TCR expression defect, a CD3 or CD247 defect may be suspected and further studies are required to identify the culprit protein (see “Protein identification in patients’ lymphocytes”), as indicated in the diagnostic flow chart (
Figure 3, bottom). The following TCR expression hierarchy may be useful for the diagnosis of TCRID due to the lack of the indicated chains: CD3ɛ=CD3δ=TCRα≥CD247>CD3γ (our experience). Also, studying NK cells can be informative, as CD247 deficiency affects the expression of NK surface receptors such as CD16 or NKp30 (Reyburn, unpublished data). It is important to note that in contrast to αβ T cells, the range of surface TCR levels in γδ T cells in different individuals is quite heterogeneous (
Garcillán et al. 2014), a fact that must be taken into account when analyzing the overlays of γδ T cells.
Lastly, it is advisable to grow T cells using IL-2 plus allogeneic feeder cells or immortalize them with Herpesvirus saimiri (HVS) or Human T-cell leukemia virus type 1 (HTLV-1), (see “Functional studies” and “Cellular models”), so that the detected TCR expression defects can be ascertained and further analyzed.
Protein identification in patients’ lymphocytes
In the past, immunoprecipitation (
Perez-Aciego et al. 1991) or Western blot techniques (
Gil et al. 2011) had to be used for the identification of protein defects. But these approaches require large amounts of T cells, which are difficult to obtain when samples are scarce (as is often the case with infant patients), and even more difficult in cases with severe T-cell lymphopenia. Generation of T-cell lines can be a solution, but they take precious time, thereby delaying diagnosis. CD3 chain-specific antibodies for intracellular staining have become available lately, and in our hands simplified the diagnosis of TCRID, which can now be done rapidly.
Table 3 (intracellular) lists such antibodies and
Figure 3 (top right) shows a recent example of their use. Once the culprit protein is defined, genetic validation is indispensable to establish the molecular basis of the protein defect.
In summary, excellent reagents are now available to diagnose TCRID defects in a record time, even when very few T cells are present.
Mutation analysis
A mutation database has been established that contains most described mutations in the genes encoding for TCR chains (
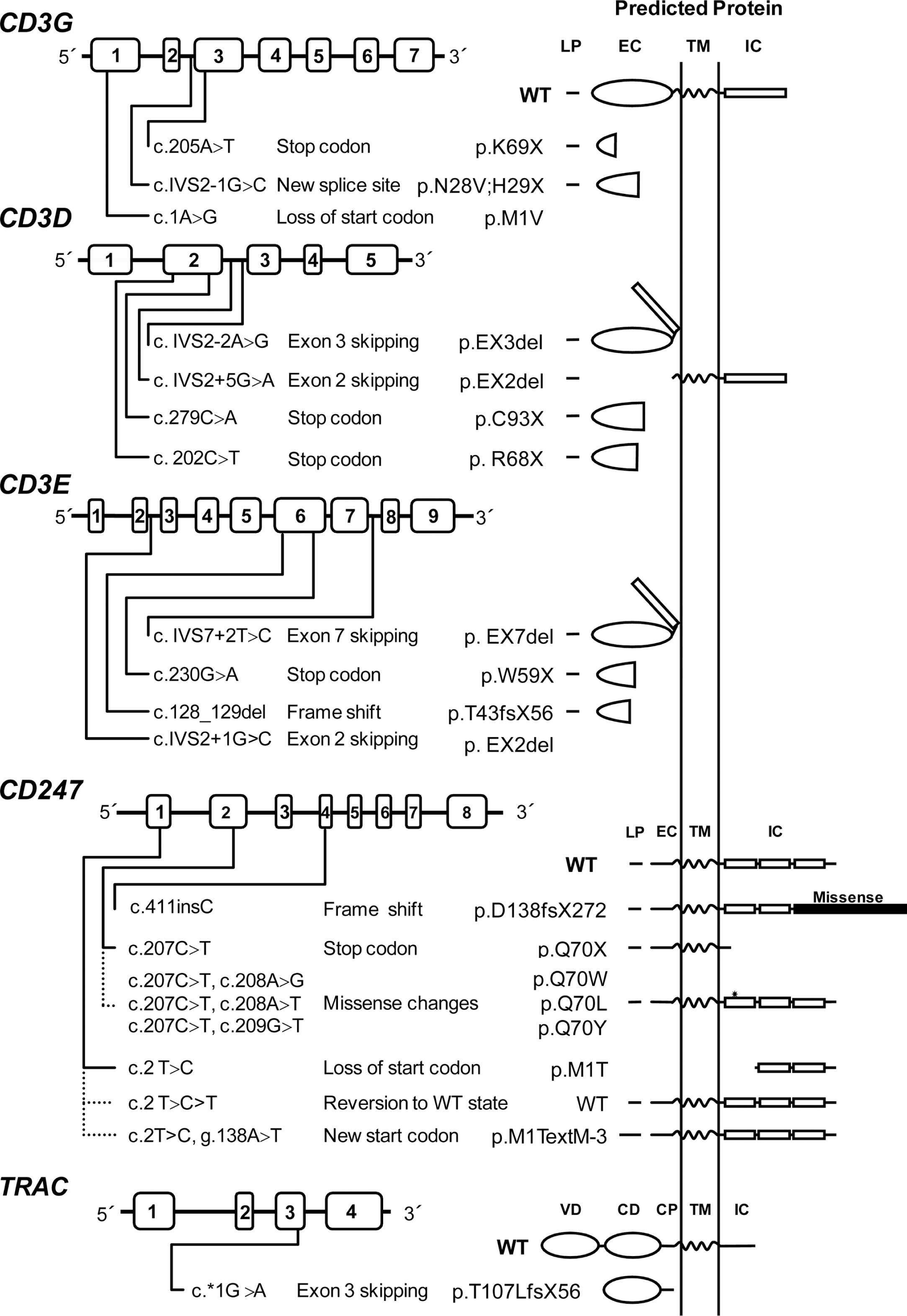
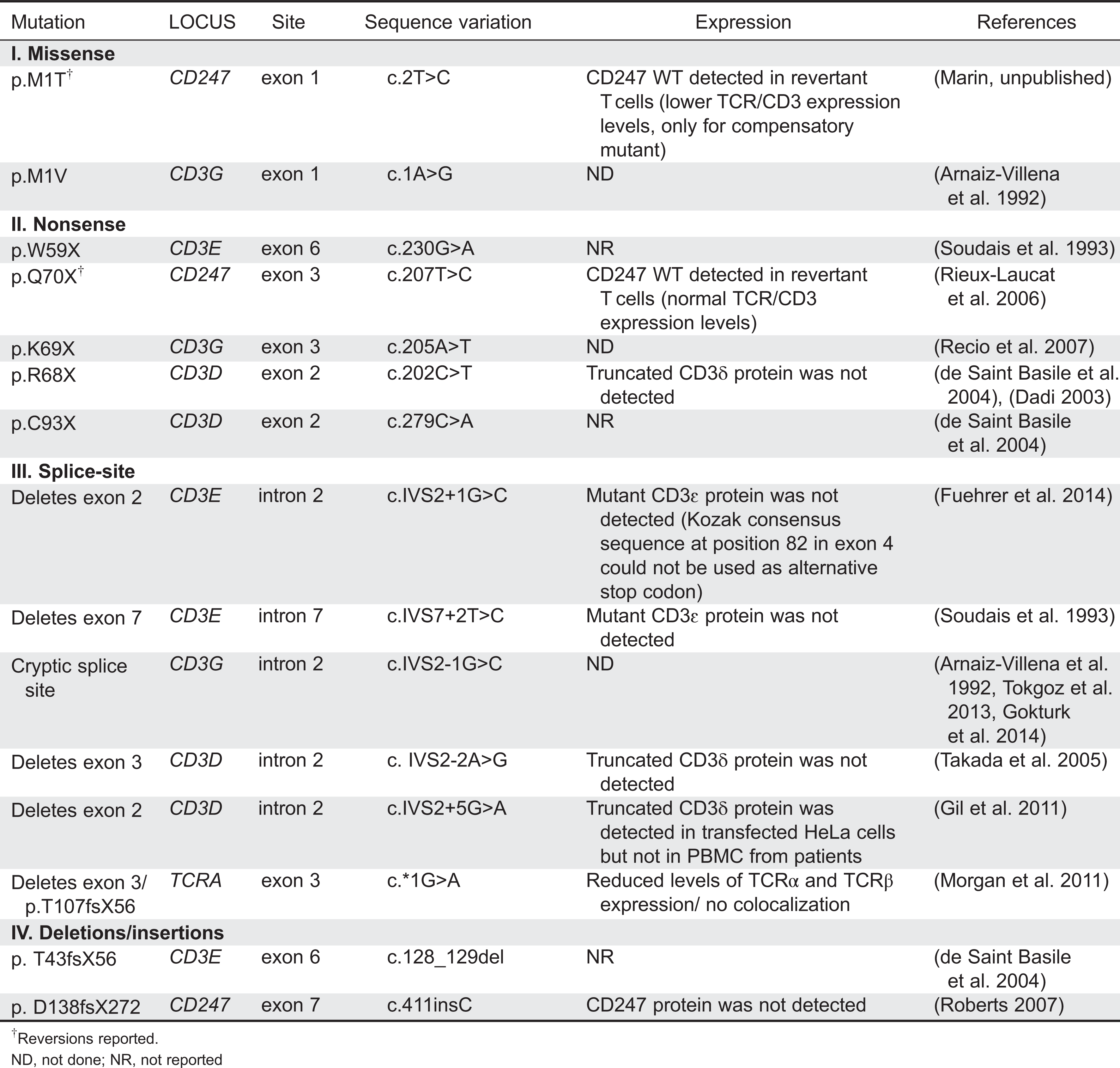
Piirilä et al. 2006). Mutations causing TCRID include missense, nonsense, and splice-site mutations as well as less common genomic deletions and insertions (
Table 4).
Figure 4 illustrates their presumed protein products that, when truncated, are often unstable.
Most mutations drastically affect the expression of the protein (complete defects), whereas in some leaky splice-site mutations, small amounts of normal transcripts were produced, allowing for the expression of some normal protein and thus causing a partial phenotype. This is the case for CD3δ and CD3ɛ TCRID.
Potentially truncated proteins have also been reported and were likely unstable, as they were undetectable in T cells. Indeed, the in-frame deletion of exon 7 predicted a mutant CD3ɛ protein that was not found even with very sensitive techniques (
Thoenes et al. 1992). Also, a predicted mutant CD3δ protein selectively lacking the extracellular domain encoded by exon 2 was undetectable in peripheral blood mononuclear cells (PBMC) from the patient or carriers, although it could be shown in transfected non-T cells (
Gil et al. 2011). This may be mutation specific, as shown recently in a new splicing mutation in
CD3E intron 2, which was predicted to delete exon 2 and thus the start codon. As a consequence, no protein was detected by Western blot either in carrier PBMC or in transfected non-T cells (
Fuehrer et al. 2014). In TCRα deficiency, a c.*1G>A mutation resulted in an aberrant transcript joining exon 2 to the normally untranslated exon 4. In the predicted translation product, the 35 C-terminal amino acids would be replaced by 56 amino acids encoded by exon 4. This would result in partial loss of the connecting peptide domain and abolition of the transmembrane and cytoplasmic domains of the TCRα chain. In PBMC, no TCRα or TCRβ proteins were detected by Western blotting with specific antibodies, but minute amounts of both chains were detectable by immunofluorescence microscopy using a different set of antibodies. The authors concluded that the residual predicted truncated TCRα failed to bind normally to TCRβ, because in the latter experiment the TCR chains did not colocalize. This was later confirmed in transfected HeLa cells (
Morgan et al. 2011).
Nonsense mutations are clearly deleterious and frequently associate with severe TCRID immunophenotypes, except for complete CD3γ and partial CD3ɛ deficiencies. In CD3δ deficiency, a microarray analysis in thymocytes revealed more than 2-fold lower specific transcripts, but no CD3δ protein was detected by Western blot analysis of the same lysates or by thymus immunohistochemistry (
Dadi et al. 2003)
.Somatic mosaicism caused by reversions can ease otherwise severe clinical phenotypes. Reversions have been reported in several disorders, including dermatological, metabolic, and immunological disorders such as ADA deficiency, X-linked SCID, and Wiskott–Aldrich syndrome (
Hirschhorn 2003;
Wada and Candotti 2008).
Reversion of some T cells to carrier TCR expression levels has been shown to be the consequence of
CD247 germ-line mutations when studied (
Figure 4). However, TCR signaling after stimulation with phytohemagglutinin (PHA) or anti-CD3 of revertant T cells can be impaired (
Rieux-Laucat et al. 2006) or restored to carrier levels (Marin, unpublished data) for reasons yet to be defined. Interestingly, no reversions have been reported in other TCRID, suggesting that
CD247 is more prone to somatic mutations than the
CD3 or
TRAC genes.
CD3GDE haplotype analysis using polymorphic markers can be useful (
i) to rule out CD3 mutations, if segregation does not fit the pedigree; (
ii) to establish the existence of founder alleles even when families are unaware of such connections, as we have reported for
CD3G in Turkey (
Recio et al. 2007) and for
CD3D in Ecuador (
Gil et al. 2011); and (
iii) for carrier detection and (or) prenatal diagnosis, because recombination within the
CD3 gene complex is rare.
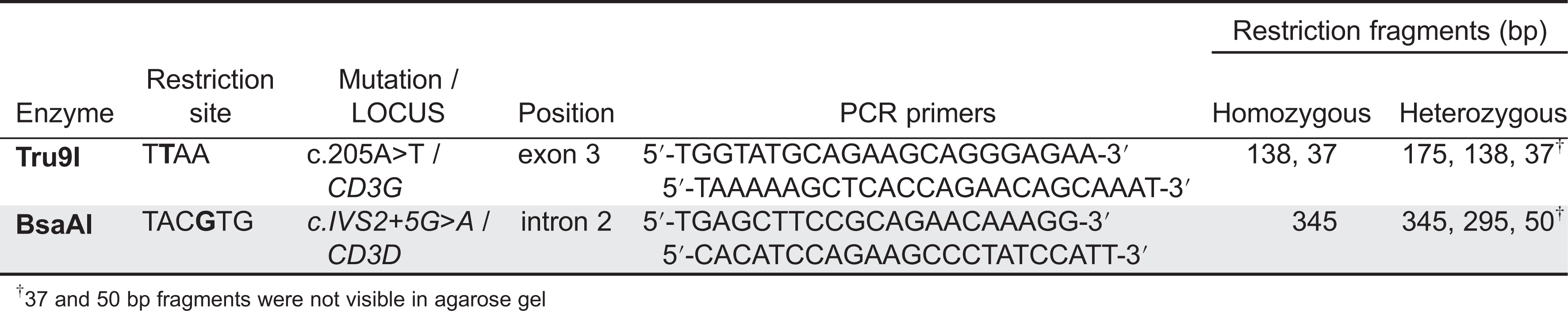
Screening tests specific for some founder
CD3G and
CD3D mutations using TaqI and BsaAI (
Table 5) have been developed for several Turkish and Ecuadorian families, respectively, which help to reach quick diagnoses and segregation data for genetic counselling (
Recio et al. 2007;
Gil et al. 2011). Therefore, in Turkish or Ecuadorian patients with an immunophenotype suggestive of CD3γ or CD3δ deficiency, regardless of the accompanying spectrum of clinical symptoms, screening for such mutations should be considered in their molecular diagnosis.